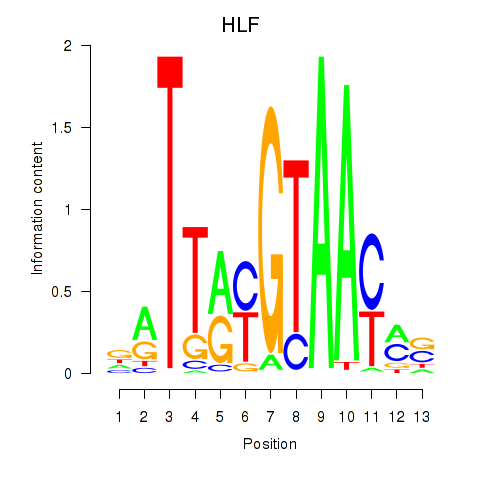
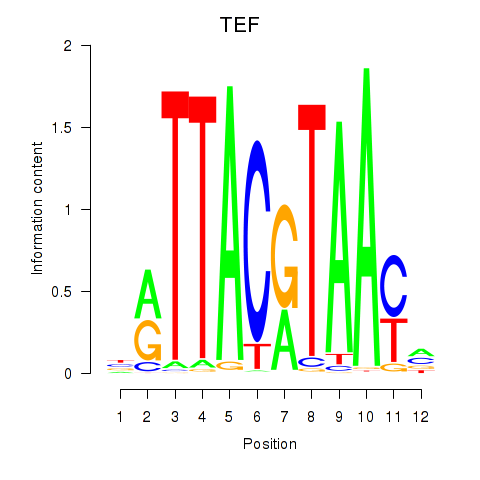
|
chr6_-_46491431
Show fit
|
0.18 |
ENST00000371374.6
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_+_17302854
Show fit
|
0.12 |
ENST00000594999.1
|
MRPL34
|
mitochondrial ribosomal protein L34
|
|
chr1_-_156751654
Show fit
|
0.10 |
ENST00000357325.10
|
HDGF
|
heparin binding growth factor
|
|
chr1_-_156751597
Show fit
|
0.10 |
ENST00000537739.5
|
HDGF
|
heparin binding growth factor
|
|
chr2_-_202238462
Show fit
|
0.09 |
ENST00000409205.5
|
SUMO1
|
small ubiquitin like modifier 1
|
|
chr14_+_75428011
Show fit
|
0.09 |
ENST00000651602.1
ENST00000559060.5
|
JDP2
|
Jun dimerization protein 2
|
|
chr3_-_190862688
Show fit
|
0.09 |
ENST00000442080.6
|
GMNC
|
geminin coiled-coil domain containing
|
|
chr20_+_63739751
Show fit
|
0.08 |
ENST00000266077.5
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr21_+_25639251
Show fit
|
0.08 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2
|
|
chr17_-_43545636
Show fit
|
0.08 |
ENST00000393664.6
|
ETV4
|
ETS variant transcription factor 4
|
|
chr2_-_212124901
Show fit
|
0.08 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
chr17_-_43545891
Show fit
|
0.08 |
ENST00000591713.5
ENST00000538265.5
|
ETV4
|
ETS variant transcription factor 4
|
|
chr7_-_141702151
Show fit
|
0.08 |
ENST00000536163.6
|
DENND11
|
DENN domain containing 11
|
|
chr17_-_43545707
Show fit
|
0.07 |
ENST00000545089.5
|
ETV4
|
ETS variant transcription factor 4
|
|
chr17_-_43546323
Show fit
|
0.07 |
ENST00000545954.5
ENST00000319349.10
|
ETV4
|
ETS variant transcription factor 4
|
|
chr17_-_782253
Show fit
|
0.07 |
ENST00000628529.2
ENST00000625892.1
ENST00000301328.9
ENST00000576419.1
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr1_-_243850070
Show fit
|
0.07 |
ENST00000366539.6
ENST00000672578.1
|
AKT3
|
AKT serine/threonine kinase 3
|
|
chr16_-_705726
Show fit
|
0.07 |
ENST00000397621.6
ENST00000324361.9
|
FBXL16
|
F-box and leucine rich repeat protein 16
|
|
chr22_-_30471986
Show fit
|
0.07 |
ENST00000401751.5
ENST00000402286.5
ENST00000403066.5
ENST00000215812.9
|
SEC14L3
|
SEC14 like lipid binding 3
|
|
chr21_+_25639272
Show fit
|
0.07 |
ENST00000400532.5
ENST00000312957.9
|
JAM2
|
junctional adhesion molecule 2
|
|
chr5_-_111756245
Show fit
|
0.07 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein
|
|
chr17_-_782317
Show fit
|
0.07 |
ENST00000301329.10
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr8_+_67952028
Show fit
|
0.06 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2
|
|
chr14_-_21537206
Show fit
|
0.06 |
ENST00000614342.1
|
SALL2
|
spalt like transcription factor 2
|
|
chr16_+_70646536
Show fit
|
0.06 |
ENST00000288098.6
|
IL34
|
interleukin 34
|
|
chr11_-_62545629
Show fit
|
0.06 |
ENST00000528508.5
ENST00000533365.5
|
AHNAK
|
AHNAK nucleoprotein
|
|
chr3_+_148730100
Show fit
|
0.06 |
ENST00000474935.5
ENST00000475347.5
ENST00000461609.1
|
AGTR1
|
angiotensin II receptor type 1
|
|
chr17_-_19748285
Show fit
|
0.06 |
ENST00000570414.1
ENST00000225740.11
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr1_-_161118024
Show fit
|
0.06 |
ENST00000368010.4
|
PFDN2
|
prefoldin subunit 2
|
|
chr1_+_65264694
Show fit
|
0.06 |
ENST00000263441.11
ENST00000395325.7
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6
|
|
chr3_-_48088800
Show fit
|
0.06 |
ENST00000423088.5
|
MAP4
|
microtubule associated protein 4
|
|
chr17_-_19748341
Show fit
|
0.06 |
ENST00000395555.7
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr21_-_6467509
Show fit
|
0.05 |
ENST00000624406.3
ENST00000398168.5
ENST00000624934.3
|
CBSL
|
cystathionine beta-synthase like
|
|
chr3_-_48089203
Show fit
|
0.05 |
ENST00000468075.2
ENST00000360240.10
|
MAP4
|
microtubule associated protein 4
|
|
chr3_+_42091316
Show fit
|
0.05 |
ENST00000327628.10
|
TRAK1
|
trafficking kinesin protein 1
|
|
chr18_+_35041387
Show fit
|
0.05 |
ENST00000538170.6
ENST00000300249.10
ENST00000588910.5
|
MAPRE2
|
microtubule associated protein RP/EB family member 2
|
|
chr15_-_82952683
Show fit
|
0.05 |
ENST00000450735.7
ENST00000304231.12
|
HOMER2
|
homer scaffold protein 2
|
|
chr21_+_43719095
Show fit
|
0.05 |
ENST00000468090.5
ENST00000291565.9
|
PDXK
|
pyridoxal kinase
|
|
chr11_-_106077401
Show fit
|
0.05 |
ENST00000526793.5
|
KBTBD3
|
kelch repeat and BTB domain containing 3
|
|
chr8_-_101205455
Show fit
|
0.05 |
ENST00000520984.5
|
ZNF706
|
zinc finger protein 706
|
|
chr10_-_80289647
Show fit
|
0.05 |
ENST00000372213.8
|
MAT1A
|
methionine adenosyltransferase 1A
|
|
chr1_-_72100930
Show fit
|
0.05 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1
|
|
chr16_-_53503192
Show fit
|
0.05 |
ENST00000568596.5
ENST00000394657.12
ENST00000570004.5
ENST00000564497.1
ENST00000300245.8
|
AKTIP
|
AKT interacting protein
|
|
chr17_-_81937221
Show fit
|
0.04 |
ENST00000402252.6
ENST00000583564.5
ENST00000585244.1
ENST00000337943.9
ENST00000579698.5
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr1_+_40373697
Show fit
|
0.04 |
ENST00000372718.8
|
SMAP2
|
small ArfGAP2
|
|
chr11_-_106077313
Show fit
|
0.04 |
ENST00000531837.2
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB domain containing 3
|
|
chr5_-_179623098
Show fit
|
0.04 |
ENST00000630639.1
ENST00000329433.10
ENST00000510411.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr17_-_81937277
Show fit
|
0.04 |
ENST00000405481.8
ENST00000329875.13
ENST00000585215.5
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr17_-_81937320
Show fit
|
0.04 |
ENST00000577624.5
ENST00000403172.8
ENST00000619204.4
ENST00000629768.2
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr6_-_132734692
Show fit
|
0.04 |
ENST00000509351.5
ENST00000417437.6
ENST00000423615.6
ENST00000427187.6
ENST00000414302.7
ENST00000367927.9
ENST00000450865.2
|
VNN3
|
vanin 3
|
|
chr17_-_75182536
Show fit
|
0.04 |
ENST00000578238.2
|
SUMO2
|
small ubiquitin like modifier 2
|
|
chr8_-_27611424
Show fit
|
0.04 |
ENST00000405140.7
|
CLU
|
clusterin
|
|
chr15_+_67125707
Show fit
|
0.04 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr9_+_98943898
Show fit
|
0.04 |
ENST00000375001.8
|
COL15A1
|
collagen type XV alpha 1 chain
|
|
chr17_-_44067585
Show fit
|
0.03 |
ENST00000591247.6
|
LSM12
|
LSM12 homolog
|
|
chr11_-_77474087
Show fit
|
0.03 |
ENST00000356341.8
|
PAK1
|
p21 (RAC1) activated kinase 1
|
|
chr1_+_115029823
Show fit
|
0.03 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta
|
|
chr6_-_30075767
Show fit
|
0.03 |
ENST00000244360.8
ENST00000376751.8
|
RNF39
|
ring finger protein 39
|
|
chr12_-_120529140
Show fit
|
0.03 |
ENST00000552443.5
ENST00000547736.1
ENST00000288532.11
ENST00000445328.6
ENST00000547943.5
|
COQ5
|
coenzyme Q5, methyltransferase
|
|
chr2_-_166375969
Show fit
|
0.03 |
ENST00000454569.6
ENST00000409672.5
|
SCN9A
|
sodium voltage-gated channel alpha subunit 9
|
|
chr14_+_101809795
Show fit
|
0.03 |
ENST00000350249.7
ENST00000557621.5
ENST00000556946.1
|
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma
|
|
chr3_+_141384790
Show fit
|
0.03 |
ENST00000507722.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr2_-_166375901
Show fit
|
0.03 |
ENST00000303354.11
ENST00000645907.1
ENST00000642356.2
ENST00000452182.2
|
SCN9A
|
sodium voltage-gated channel alpha subunit 9
|
|
chr13_-_33205997
Show fit
|
0.03 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13
|
|
chr8_-_27611325
Show fit
|
0.03 |
ENST00000523500.5
|
CLU
|
clusterin
|
|
chr21_-_43659460
Show fit
|
0.03 |
ENST00000443485.1
ENST00000291560.7
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr17_+_41819201
Show fit
|
0.03 |
ENST00000455106.1
|
FKBP10
|
FKBP prolyl isomerase 10
|
|
chr1_+_61203496
Show fit
|
0.03 |
ENST00000663597.1
|
NFIA
|
nuclear factor I A
|
|
chr12_+_57230301
Show fit
|
0.03 |
ENST00000553474.5
|
SHMT2
|
serine hydroxymethyltransferase 2
|
|
chr19_-_40882226
Show fit
|
0.03 |
ENST00000301146.9
|
CYP2A7
|
cytochrome P450 family 2 subfamily A member 7
|
|
chr7_+_44606563
Show fit
|
0.03 |
ENST00000439616.6
|
OGDH
|
oxoglutarate dehydrogenase
|
|
chr11_-_77474041
Show fit
|
0.03 |
ENST00000278568.8
|
PAK1
|
p21 (RAC1) activated kinase 1
|
|
chr1_+_212565334
Show fit
|
0.03 |
ENST00000366981.8
ENST00000366987.6
|
ATF3
|
activating transcription factor 3
|
|
chr21_+_39452077
Show fit
|
0.03 |
ENST00000452550.5
|
SH3BGR
|
SH3 domain binding glutamate rich protein
|
|
chr3_+_155080307
Show fit
|
0.03 |
ENST00000360490.7
|
MME
|
membrane metalloendopeptidase
|
|
chr6_+_72216442
Show fit
|
0.02 |
ENST00000425662.6
ENST00000453976.6
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_-_44848021
Show fit
|
0.02 |
ENST00000349299.7
ENST00000521529.5
ENST00000350771.7
ENST00000222690.10
ENST00000381124.9
ENST00000437072.5
ENST00000446531.1
ENST00000308153.9
|
H2AZ2
|
H2A.Z variant histone 2
|
|
chr13_-_37598750
Show fit
|
0.02 |
ENST00000379743.8
ENST00000379742.4
ENST00000379749.8
ENST00000379747.9
ENST00000541179.5
ENST00000541481.5
|
POSTN
|
periostin
|
|
chr10_-_14330879
Show fit
|
0.02 |
ENST00000357447.7
|
FRMD4A
|
FERM domain containing 4A
|
|
chr2_+_190343561
Show fit
|
0.02 |
ENST00000322522.8
ENST00000392329.7
ENST00000430311.5
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chr3_-_48088824
Show fit
|
0.02 |
ENST00000439356.2
ENST00000395734.7
ENST00000426837.6
|
MAP4
|
microtubule associated protein 4
|
|
chr12_+_93569814
Show fit
|
0.02 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr15_-_65792090
Show fit
|
0.02 |
ENST00000431932.6
|
DENND4A
|
DENN domain containing 4A
|
|
chr19_+_51573171
Show fit
|
0.02 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175
|
|
chr9_+_98943705
Show fit
|
0.02 |
ENST00000610452.1
|
COL15A1
|
collagen type XV alpha 1 chain
|
|
chr5_-_147906530
Show fit
|
0.02 |
ENST00000318315.5
ENST00000515291.1
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr7_-_31340678
Show fit
|
0.02 |
ENST00000297142.4
|
NEUROD6
|
neuronal differentiation 6
|
|
chr1_-_101025763
Show fit
|
0.02 |
ENST00000342173.11
ENST00000488176.1
ENST00000370109.8
|
DPH5
|
diphthamide biosynthesis 5
|
|
chr1_+_113905156
Show fit
|
0.02 |
ENST00000650596.1
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr6_+_96562548
Show fit
|
0.02 |
ENST00000541107.5
ENST00000326771.2
|
FHL5
|
four and a half LIM domains 5
|
|
chr11_+_62728465
Show fit
|
0.02 |
ENST00000316461.9
|
TTC9C
|
tetratricopeptide repeat domain 9C
|
|
chr19_-_45886120
Show fit
|
0.02 |
ENST00000302165.5
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr20_+_41028814
Show fit
|
0.02 |
ENST00000361337.3
|
TOP1
|
DNA topoisomerase I
|
|
chr7_+_135662467
Show fit
|
0.02 |
ENST00000507606.3
|
STMP1
|
short transmembrane mitochondrial protein 1
|
|
chr20_+_38581186
Show fit
|
0.02 |
ENST00000373348.4
ENST00000537425.2
ENST00000416116.2
|
ADIG
|
adipogenin
|
|
chr4_+_26343156
Show fit
|
0.02 |
ENST00000680928.1
ENST00000681260.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr9_+_107283256
Show fit
|
0.02 |
ENST00000358015.8
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein
|
|
chr7_+_44606578
Show fit
|
0.02 |
ENST00000443864.6
ENST00000447398.5
ENST00000449767.5
ENST00000419661.5
|
OGDH
|
oxoglutarate dehydrogenase
|
|
chr1_-_161238163
Show fit
|
0.02 |
ENST00000367982.8
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3
|
|
chr2_+_178320228
Show fit
|
0.02 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6
|
|
chr7_+_44606619
Show fit
|
0.02 |
ENST00000222673.6
ENST00000444676.5
ENST00000631326.2
|
OGDH
|
oxoglutarate dehydrogenase
|
|
chr3_+_32106612
Show fit
|
0.02 |
ENST00000282541.10
ENST00000425459.5
ENST00000431009.1
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1 like
|
|
chr17_-_75855204
Show fit
|
0.02 |
ENST00000589642.5
ENST00000593002.1
ENST00000590221.5
ENST00000587374.5
ENST00000585462.5
ENST00000254806.8
ENST00000433525.6
ENST00000626827.2
|
WBP2
|
WW domain binding protein 2
|
|
chr11_-_14972273
Show fit
|
0.02 |
ENST00000396372.2
ENST00000361010.7
ENST00000331587.9
|
CALCA
|
calcitonin related polypeptide alpha
|
|
chrX_+_41447322
Show fit
|
0.02 |
ENST00000378220.2
ENST00000342595.2
|
NYX
|
nyctalopin
|
|
chr13_-_49401497
Show fit
|
0.02 |
ENST00000457041.5
ENST00000355854.8
|
CAB39L
|
calcium binding protein 39 like
|
|
chr16_-_84618067
Show fit
|
0.02 |
ENST00000262428.5
|
COTL1
|
coactosin like F-actin binding protein 1
|
|
chr21_+_34364003
Show fit
|
0.02 |
ENST00000290310.4
|
KCNE2
|
potassium voltage-gated channel subfamily E regulatory subunit 2
|
|
chr12_+_57230336
Show fit
|
0.02 |
ENST00000555773.5
ENST00000554975.5
ENST00000449049.7
|
SHMT2
|
serine hydroxymethyltransferase 2
|
|
chr14_-_20454741
Show fit
|
0.02 |
ENST00000206542.9
ENST00000553640.3
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr3_+_157436842
Show fit
|
0.02 |
ENST00000295927.4
|
PTX3
|
pentraxin 3
|
|
chr7_-_73738792
Show fit
|
0.02 |
ENST00000222800.8
ENST00000458339.6
|
ABHD11
|
abhydrolase domain containing 11
|
|
chr16_-_84618041
Show fit
|
0.02 |
ENST00000564057.1
|
COTL1
|
coactosin like F-actin binding protein 1
|
|
chr8_-_101205240
Show fit
|
0.02 |
ENST00000520347.5
ENST00000523922.1
|
ZNF706
|
zinc finger protein 706
|
|
chr12_-_52680398
Show fit
|
0.02 |
ENST00000252244.3
|
KRT1
|
keratin 1
|
|
chr8_-_101206064
Show fit
|
0.02 |
ENST00000518336.5
ENST00000520454.1
|
ZNF706
|
zinc finger protein 706
|
|
chr17_-_43907467
Show fit
|
0.02 |
ENST00000520406.5
ENST00000518478.1
ENST00000522172.5
ENST00000461854.5
ENST00000521178.5
ENST00000269095.9
ENST00000520305.5
ENST00000523501.5
ENST00000520241.5
|
MPP2
|
membrane palmitoylated protein 2
|
|
chr5_-_59586393
Show fit
|
0.02 |
ENST00000505453.1
ENST00000360047.9
|
PDE4D
|
phosphodiesterase 4D
|
|
chr11_-_35360050
Show fit
|
0.02 |
ENST00000644868.1
ENST00000643454.1
ENST00000646080.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr2_+_9961165
Show fit
|
0.02 |
ENST00000405379.6
|
GRHL1
|
grainyhead like transcription factor 1
|
|
chr7_+_20647388
Show fit
|
0.02 |
ENST00000258738.10
|
ABCB5
|
ATP binding cassette subfamily B member 5
|
|
chr4_+_41613476
Show fit
|
0.02 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr11_+_124115404
Show fit
|
0.02 |
ENST00000361352.9
ENST00000449321.5
ENST00000392748.5
ENST00000392744.4
ENST00000456829.7
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr9_+_32552305
Show fit
|
0.02 |
ENST00000451672.2
ENST00000644531.1
|
SMIM27
|
small integral membrane protein 27
|
|
chr5_-_43557689
Show fit
|
0.02 |
ENST00000338972.8
ENST00000511321.5
ENST00000515338.1
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chr10_+_18340699
Show fit
|
0.01 |
ENST00000377329.10
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr11_+_106077638
Show fit
|
0.01 |
ENST00000278618.9
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase
|
|
chr16_-_70289480
Show fit
|
0.01 |
ENST00000261772.13
ENST00000675953.1
ENST00000675691.1
ENST00000675133.1
ENST00000674512.1
ENST00000675045.1
ENST00000675853.1
ENST00000565361.3
ENST00000674963.1
ENST00000674691.1
|
AARS1
|
alanyl-tRNA synthetase 1
|
|
chr14_+_60249191
Show fit
|
0.01 |
ENST00000395076.9
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A
|
|
chr8_-_144060681
Show fit
|
0.01 |
ENST00000618853.5
|
OPLAH
|
5-oxoprolinase, ATP-hydrolysing
|
|
chr12_-_101407727
Show fit
|
0.01 |
ENST00000539055.5
ENST00000551688.1
ENST00000551671.5
ENST00000261636.13
|
ARL1
|
ADP ribosylation factor like GTPase 1
|
|
chr19_+_33373694
Show fit
|
0.01 |
ENST00000284000.9
|
CEBPG
|
CCAAT enhancer binding protein gamma
|
|
chr16_+_70299156
Show fit
|
0.01 |
ENST00000393657.6
ENST00000288071.11
ENST00000355992.7
ENST00000567706.1
ENST00000443119.7
|
DDX19B
ENSG00000260537.2
|
DEAD-box helicase 19B
novel protein, DDX19B and DDX19A readthrough
|
|
chr9_-_125484490
Show fit
|
0.01 |
ENST00000444226.1
|
MAPKAP1
|
MAPK associated protein 1
|
|
chr9_-_127950716
Show fit
|
0.01 |
ENST00000373084.8
|
FAM102A
|
family with sequence similarity 102 member A
|
|
chr8_-_101204697
Show fit
|
0.01 |
ENST00000517844.5
|
ZNF706
|
zinc finger protein 706
|
|
chr1_-_161238085
Show fit
|
0.01 |
ENST00000512372.5
ENST00000437437.6
ENST00000412844.6
ENST00000428574.6
ENST00000442691.6
ENST00000505005.5
ENST00000508740.5
ENST00000367981.7
ENST00000502985.5
ENST00000504010.5
ENST00000508387.5
ENST00000511676.5
ENST00000511748.5
ENST00000511944.5
ENST00000515621.5
ENST00000367984.8
ENST00000367985.7
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3
|
|
chr5_-_150758497
Show fit
|
0.01 |
ENST00000521533.1
ENST00000424236.5
|
DCTN4
|
dynactin subunit 4
|
|
chr8_-_101205561
Show fit
|
0.01 |
ENST00000519744.5
ENST00000311212.9
ENST00000521272.5
ENST00000519882.5
|
ZNF706
|
zinc finger protein 706
|
|
chr7_+_23106267
Show fit
|
0.01 |
ENST00000409689.5
ENST00000410047.1
|
KLHL7
|
kelch like family member 7
|
|
chr10_+_92291155
Show fit
|
0.01 |
ENST00000358935.3
|
MARCHF5
|
membrane associated ring-CH-type finger 5
|
|
chr17_+_782340
Show fit
|
0.01 |
ENST00000304478.9
|
MRM3
|
mitochondrial rRNA methyltransferase 3
|
|
chr8_+_106726012
Show fit
|
0.01 |
ENST00000449762.6
ENST00000297447.10
|
OXR1
|
oxidation resistance 1
|
|
chr20_+_16748358
Show fit
|
0.01 |
ENST00000246081.3
|
OTOR
|
otoraplin
|
|
chr1_-_167914089
Show fit
|
0.01 |
ENST00000476818.2
ENST00000367848.1
ENST00000367851.9
ENST00000545172.5
|
ADCY10
|
adenylate cyclase 10
|
|
chr11_-_124313949
Show fit
|
0.01 |
ENST00000641015.1
|
OR8D1
|
olfactory receptor family 8 subfamily D member 1
|
|
chr4_+_186266183
Show fit
|
0.01 |
ENST00000403665.7
ENST00000492972.6
ENST00000264692.8
|
F11
|
coagulation factor XI
|
|
chr14_+_101809855
Show fit
|
0.01 |
ENST00000557714.1
ENST00000445439.7
ENST00000334743.9
ENST00000557095.5
|
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma
|
|
chr4_+_69280472
Show fit
|
0.01 |
ENST00000335568.10
ENST00000511240.1
|
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28
|
|
chr19_+_33374312
Show fit
|
0.01 |
ENST00000585933.2
|
CEBPG
|
CCAAT enhancer binding protein gamma
|
|
chr21_+_43659528
Show fit
|
0.01 |
ENST00000340648.6
|
RRP1B
|
ribosomal RNA processing 1B
|
|
chr1_-_161238196
Show fit
|
0.01 |
ENST00000367983.9
ENST00000506209.5
ENST00000367980.6
ENST00000628566.2
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3
|
|
chr15_+_49621004
Show fit
|
0.01 |
ENST00000403028.8
ENST00000558653.5
ENST00000559164.5
ENST00000560632.5
ENST00000559405.5
ENST00000251250.7
|
DTWD1
|
DTW domain containing 1
|
|
chr12_+_57230086
Show fit
|
0.01 |
ENST00000414700.7
ENST00000557703.5
|
SHMT2
|
serine hydroxymethyltransferase 2
|
|
chrX_+_9463272
Show fit
|
0.01 |
ENST00000407597.7
ENST00000380961.5
ENST00000424279.6
|
TBL1X
|
transducin beta like 1 X-linked
|
|
chr6_+_96924614
Show fit
|
0.01 |
ENST00000536676.5
ENST00000539200.5
ENST00000544166.5
|
KLHL32
|
kelch like family member 32
|
|
chr1_-_226309198
Show fit
|
0.01 |
ENST00000481685.1
|
LIN9
|
lin-9 DREAM MuvB core complex component
|
|
chr18_+_13824149
Show fit
|
0.01 |
ENST00000589410.2
|
MC5R
|
melanocortin 5 receptor
|
|
chr14_+_92794297
Show fit
|
0.01 |
ENST00000163416.7
|
GOLGA5
|
golgin A5
|
|
chr12_-_75209814
Show fit
|
0.01 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2
|
|
chr1_+_11189347
Show fit
|
0.01 |
ENST00000376819.4
|
ANGPTL7
|
angiopoietin like 7
|
|
chr12_-_75209701
Show fit
|
0.01 |
ENST00000350228.6
ENST00000298972.5
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2
|
|
chr8_+_24294107
Show fit
|
0.01 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr17_-_75182949
Show fit
|
0.01 |
ENST00000314523.7
ENST00000420826.7
|
SUMO2
|
small ubiquitin like modifier 2
|
|
chr6_+_96924720
Show fit
|
0.01 |
ENST00000369261.9
|
KLHL32
|
kelch like family member 32
|
|
chr22_-_49827512
Show fit
|
0.01 |
ENST00000404760.5
|
BRD1
|
bromodomain containing 1
|
|
chr16_+_72054477
Show fit
|
0.01 |
ENST00000355906.10
ENST00000570083.5
ENST00000228226.12
ENST00000398131.6
ENST00000569639.5
ENST00000564499.5
ENST00000357763.8
ENST00000613898.1
ENST00000562526.5
ENST00000565574.5
ENST00000568417.6
|
HP
|
haptoglobin
|
|
chr4_-_73223082
Show fit
|
0.01 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr10_-_102451536
Show fit
|
0.01 |
ENST00000625129.1
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr19_-_6459735
Show fit
|
0.01 |
ENST00000334510.9
ENST00000301454.9
|
SLC25A23
|
solute carrier family 25 member 23
|
|
chr3_+_159069252
Show fit
|
0.01 |
ENST00000640015.1
ENST00000476809.7
ENST00000485419.7
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough
|
|
chr14_-_67359769
Show fit
|
0.01 |
ENST00000555431.5
ENST00000554236.5
ENST00000555474.5
ENST00000216442.12
|
ATP6V1D
|
ATPase H+ transporting V1 subunit D
|
|
chr3_-_99850976
Show fit
|
0.01 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr8_+_49911396
Show fit
|
0.01 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1
|
|
chr2_+_197453423
Show fit
|
0.01 |
ENST00000263960.6
|
COQ10B
|
coenzyme Q10B
|
|
chr12_+_57229694
Show fit
|
0.01 |
ENST00000557487.5
ENST00000328923.8
ENST00000555634.5
ENST00000556689.5
|
SHMT2
|
serine hydroxymethyltransferase 2
|
|
chr14_+_60249387
Show fit
|
0.01 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A
|
|
chr8_+_42541775
Show fit
|
0.01 |
ENST00000416469.6
|
SMIM19
|
small integral membrane protein 19
|
|
chr10_+_18340821
Show fit
|
0.01 |
ENST00000612743.1
ENST00000612134.4
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr6_-_31972290
Show fit
|
0.01 |
ENST00000375349.7
|
DXO
|
decapping exoribonuclease
|
|
chr8_+_49911604
Show fit
|
0.01 |
ENST00000642164.1
ENST00000644093.1
ENST00000643999.1
ENST00000647073.1
ENST00000646880.1
|
SNTG1
|
syntrophin gamma 1
|
|
chr2_-_98663464
Show fit
|
0.01 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A
|
|
chr8_-_6877928
Show fit
|
0.01 |
ENST00000297439.4
|
DEFB1
|
defensin beta 1
|
|
chr3_-_99876104
Show fit
|
0.01 |
ENST00000471562.1
ENST00000495625.2
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr19_-_41439899
Show fit
|
0.01 |
ENST00000597457.5
ENST00000589970.5
ENST00000595425.5
ENST00000438807.7
ENST00000589102.1
ENST00000221943.14
ENST00000592922.6
|
DMAC2
|
distal membrane arm assembly complex 2
|
|
chr19_+_48756067
Show fit
|
0.01 |
ENST00000222157.5
|
FGF21
|
fibroblast growth factor 21
|
|
chr17_-_21042901
Show fit
|
0.01 |
ENST00000261497.9
|
USP22
|
ubiquitin specific peptidase 22
|
|
chr2_-_166128004
Show fit
|
0.00 |
ENST00000303395.9
ENST00000674923.1
ENST00000637988.1
ENST00000635776.1
|
SCN1A
|
sodium voltage-gated channel alpha subunit 1
|
|
chr19_+_48755512
Show fit
|
0.00 |
ENST00000593756.6
|
FGF21
|
fibroblast growth factor 21
|
|
chr17_-_34363224
Show fit
|
0.00 |
ENST00000225842.4
|
CCL1
|
C-C motif chemokine ligand 1
|
|
chr9_-_34665985
Show fit
|
0.00 |
ENST00000416454.5
ENST00000544078.2
ENST00000421828.7
ENST00000423809.5
|
ENSG00000187186.15
|
novel protein
|
|
chr16_+_3458006
Show fit
|
0.00 |
ENST00000577013.6
ENST00000414063.6
ENST00000572584.2
|
NAA60
|
N-alpha-acetyltransferase 60, NatF catalytic subunit
|
|
chr1_+_44746401
Show fit
|
0.00 |
ENST00000372217.5
|
KIF2C
|
kinesin family member 2C
|
|
chr2_+_68467544
Show fit
|
0.00 |
ENST00000303795.9
|
APLF
|
aprataxin and PNKP like factor
|
|
chr11_-_72434330
Show fit
|
0.00 |
ENST00000542555.2
ENST00000535990.5
ENST00000294053.9
ENST00000538039.6
ENST00000340729.9
|
CLPB
|
caseinolytic mitochondrial matrix peptidase chaperone subunit B
|
|
chr20_+_2101807
Show fit
|
0.00 |
ENST00000381482.8
|
STK35
|
serine/threonine kinase 35
|
|
chr12_+_66189178
Show fit
|
0.00 |
ENST00000545837.1
|
IRAK3
|
interleukin 1 receptor associated kinase 3
|
|
chr3_-_197183849
Show fit
|
0.00 |
ENST00000443183.5
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr11_-_64879675
Show fit
|
0.00 |
ENST00000359393.6
ENST00000433803.1
ENST00000411683.1
|
EHD1
|
EH domain containing 1
|
|
chr16_+_3458063
Show fit
|
0.00 |
ENST00000574423.2
ENST00000576787.5
ENST00000572942.5
ENST00000576916.5
ENST00000575076.5
ENST00000572131.5
|
ENSG00000263212.2
NAA60
|
novel transcript
N-alpha-acetyltransferase 60, NatF catalytic subunit
|