Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
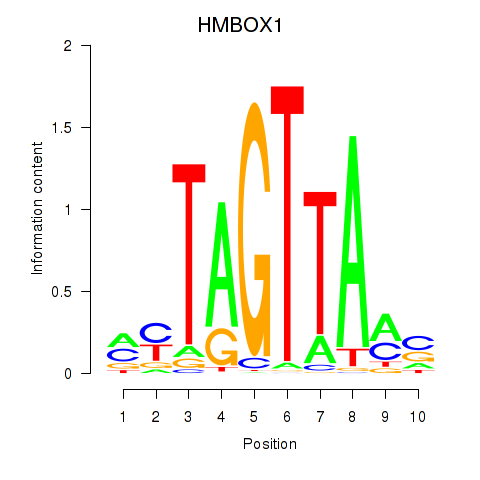
Results for HMBOX1
Z-value: 0.78
Transcription factors associated with HMBOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMBOX1
|
ENSG00000147421.18 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMBOX1 | hg38_v1_chr8_+_28890572_28890625, hg38_v1_chr8_+_28891304_28891329 | 0.19 | 6.5e-01 | Click! |
Activity profile of HMBOX1 motif
Sorted Z-values of HMBOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_117425855 | 1.09 |
ENST00000368508.7
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr6_-_117425905 | 1.00 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr5_+_93583212 | 0.85 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr12_-_55842950 | 0.72 |
ENST00000548629.5
|
MMP19
|
matrix metallopeptidase 19 |
| chr8_-_38468601 | 0.68 |
ENST00000341462.9
ENST00000683765.1 ENST00000356207.9 ENST00000326324.10 ENST00000335922.9 ENST00000532791.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr8_-_38468627 | 0.67 |
ENST00000683815.1
ENST00000684654.1 ENST00000447712.7 ENST00000397091.9 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr6_+_27147094 | 0.50 |
ENST00000377459.3
|
H2AC12
|
H2A clustered histone 12 |
| chr2_-_213151590 | 0.47 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr9_-_70414657 | 0.43 |
ENST00000377126.4
|
KLF9
|
Kruppel like factor 9 |
| chr17_+_50095285 | 0.39 |
ENST00000503614.5
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr6_+_112087576 | 0.39 |
ENST00000368656.7
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229 member B |
| chr12_-_10435940 | 0.38 |
ENST00000381901.5
ENST00000381902.7 ENST00000539033.1 |
KLRC2
ENSG00000255641.1
|
killer cell lectin like receptor C2 novel protein |
| chr19_+_49513353 | 0.37 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr3_-_18438767 | 0.35 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr13_+_23570370 | 0.35 |
ENST00000403372.6
ENST00000248484.9 |
TNFRSF19
|
TNF receptor superfamily member 19 |
| chr20_+_43558968 | 0.34 |
ENST00000647834.1
ENST00000373100.7 ENST00000648083.1 ENST00000648530.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_-_109740304 | 0.33 |
ENST00000540225.2
|
GSTM3
|
glutathione S-transferase mu 3 |
| chr4_-_158159657 | 0.33 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B |
| chr2_-_2326378 | 0.33 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr20_+_44582549 | 0.31 |
ENST00000372886.6
|
PKIG
|
cAMP-dependent protein kinase inhibitor gamma |
| chr14_+_22147988 | 0.31 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr2_-_2326210 | 0.30 |
ENST00000647755.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr10_+_87659839 | 0.30 |
ENST00000456849.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr9_-_92536031 | 0.29 |
ENST00000344604.9
ENST00000375540.5 |
ECM2
|
extracellular matrix protein 2 |
| chr5_+_173889337 | 0.29 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr16_+_85027735 | 0.28 |
ENST00000258180.7
ENST00000538274.5 |
KIAA0513
|
KIAA0513 |
| chr6_+_122717544 | 0.27 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
cAMP-dependent protein kinase inhibitor beta |
| chr2_+_46542474 | 0.26 |
ENST00000238738.9
|
RHOQ
|
ras homolog family member Q |
| chr3_-_93973933 | 0.26 |
ENST00000650591.1
|
PROS1
|
protein S |
| chr10_+_35195843 | 0.26 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chrX_+_103628959 | 0.25 |
ENST00000372625.8
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A like 1 |
| chr17_+_50095331 | 0.25 |
ENST00000503176.6
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr7_+_90709231 | 0.25 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr1_+_174875505 | 0.24 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr16_-_30585497 | 0.23 |
ENST00000567773.1
ENST00000395216.3 ENST00000470110.2 |
ZNF785
|
zinc finger protein 785 |
| chr5_+_111223905 | 0.23 |
ENST00000512453.5
|
CAMK4
|
calcium/calmodulin dependent protein kinase IV |
| chr2_-_86721122 | 0.22 |
ENST00000604011.5
|
RNF103-CHMP3
|
RNF103-CHMP3 readthrough |
| chr17_-_445939 | 0.22 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr14_+_50533026 | 0.22 |
ENST00000441560.6
|
ATL1
|
atlastin GTPase 1 |
| chr8_-_115492221 | 0.21 |
ENST00000518018.1
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr2_-_61854013 | 0.21 |
ENST00000404929.6
|
FAM161A
|
FAM161 centrosomal protein A |
| chr2_-_61854119 | 0.21 |
ENST00000405894.3
|
FAM161A
|
FAM161 centrosomal protein A |
| chrX_-_70289888 | 0.21 |
ENST00000239666.9
ENST00000374454.1 |
PDZD11
|
PDZ domain containing 11 |
| chr1_-_160282458 | 0.21 |
ENST00000485079.1
|
ENSG00000258465.8
|
novel protein |
| chr6_+_28267355 | 0.21 |
ENST00000614088.1
ENST00000619937.4 |
ZSCAN26
|
zinc finger and SCAN domain containing 26 |
| chr12_-_76486061 | 0.20 |
ENST00000548341.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr7_+_101085464 | 0.19 |
ENST00000306085.11
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr17_+_44847874 | 0.19 |
ENST00000253410.3
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr6_+_28267044 | 0.19 |
ENST00000316606.10
|
ZSCAN26
|
zinc finger and SCAN domain containing 26 |
| chr2_+_24491860 | 0.19 |
ENST00000406961.5
ENST00000405141.5 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr6_+_28267107 | 0.19 |
ENST00000621053.1
ENST00000617168.4 ENST00000421553.7 ENST00000611552.2 ENST00000623276.3 |
ENSG00000276302.1
ZSCAN26
|
novel protein zinc finger and SCAN domain containing 26 |
| chr5_+_141412979 | 0.18 |
ENST00000612503.1
ENST00000398610.3 |
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr3_-_64268161 | 0.18 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr12_+_51239278 | 0.18 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr12_-_4649043 | 0.18 |
ENST00000545990.6
ENST00000228850.6 |
AKAP3
|
A-kinase anchoring protein 3 |
| chr12_-_32896757 | 0.17 |
ENST00000070846.11
ENST00000340811.9 |
PKP2
|
plakophilin 2 |
| chr16_+_21678514 | 0.17 |
ENST00000286149.8
ENST00000388958.8 |
OTOA
|
otoancorin |
| chr5_+_141382702 | 0.17 |
ENST00000617050.1
ENST00000518325.2 |
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr4_+_73481737 | 0.17 |
ENST00000226355.5
|
AFM
|
afamin |
| chr9_+_99222258 | 0.17 |
ENST00000223641.5
|
SEC61B
|
SEC61 translocon subunit beta |
| chr19_+_48965304 | 0.17 |
ENST00000331825.11
|
FTL
|
ferritin light chain |
| chr12_-_11022620 | 0.17 |
ENST00000390673.2
|
TAS2R19
|
taste 2 receptor member 19 |
| chr17_-_3668557 | 0.16 |
ENST00000225525.4
|
TAX1BP3
|
Tax1 binding protein 3 |
| chr10_+_122560679 | 0.16 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr1_+_177170916 | 0.16 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr4_-_185956348 | 0.16 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_+_21684429 | 0.16 |
ENST00000388956.8
|
OTOA
|
otoancorin |
| chr1_+_27234612 | 0.16 |
ENST00000319394.8
ENST00000361771.7 |
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr10_+_122560751 | 0.15 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr6_-_2744126 | 0.15 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr2_+_61065863 | 0.15 |
ENST00000402291.6
|
KIAA1841
|
KIAA1841 |
| chr12_+_120534697 | 0.15 |
ENST00000675818.1
ENST00000413266.6 |
ENSG00000288623.1
RNF10
|
novel protein ring finger protein 10 |
| chr6_-_15586006 | 0.15 |
ENST00000462989.6
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr18_-_35290184 | 0.14 |
ENST00000589178.5
ENST00000592278.1 ENST00000333206.10 ENST00000592211.1 ENST00000420878.7 ENST00000610712.1 ENST00000586922.2 |
ZSCAN30
ENSG00000268573.1
|
zinc finger and SCAN domain containing 30 novel transcript |
| chr5_-_134632769 | 0.14 |
ENST00000505758.5
ENST00000439578.5 ENST00000502286.1 ENST00000402673.7 |
SAR1B
|
secretion associated Ras related GTPase 1B |
| chr7_+_134891566 | 0.14 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr22_-_19431692 | 0.13 |
ENST00000340170.8
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr22_-_36776067 | 0.13 |
ENST00000417951.6
ENST00000433985.7 ENST00000430701.5 |
IFT27
|
intraflagellar transport 27 |
| chr4_+_155903688 | 0.13 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr5_+_132369691 | 0.13 |
ENST00000245407.8
|
SLC22A5
|
solute carrier family 22 member 5 |
| chr2_+_161231078 | 0.13 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr5_+_139293939 | 0.13 |
ENST00000508689.1
ENST00000514528.5 |
MATR3
|
matrin 3 |
| chr22_-_36776147 | 0.13 |
ENST00000340630.9
|
IFT27
|
intraflagellar transport 27 |
| chr15_-_40038895 | 0.13 |
ENST00000558720.5
|
SRP14
|
signal recognition particle 14 |
| chr17_+_43025203 | 0.13 |
ENST00000587250.4
|
RND2
|
Rho family GTPase 2 |
| chr1_+_84144260 | 0.12 |
ENST00000370685.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_27771700 | 0.12 |
ENST00000379666.7
ENST00000296102.8 |
MRPL33
|
mitochondrial ribosomal protein L33 |
| chrX_+_110003095 | 0.12 |
ENST00000372073.5
ENST00000372068.7 ENST00000288381.4 |
TMEM164
|
transmembrane protein 164 |
| chr17_-_3292600 | 0.11 |
ENST00000615105.1
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1 |
| chr17_-_3668640 | 0.11 |
ENST00000611779.4
|
TAX1BP3
|
Tax1 binding protein 3 |
| chr17_-_1492660 | 0.11 |
ENST00000648651.1
|
MYO1C
|
myosin IC |
| chr14_-_50532590 | 0.11 |
ENST00000013125.9
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr22_+_23857898 | 0.11 |
ENST00000316185.8
|
SLC2A11
|
solute carrier family 2 member 11 |
| chr15_+_78873723 | 0.11 |
ENST00000559690.5
ENST00000559158.5 |
MORF4L1
|
mortality factor 4 like 1 |
| chr11_-_47426419 | 0.11 |
ENST00000298852.8
ENST00000530912.5 ENST00000619920.4 |
PSMC3
|
proteasome 26S subunit, ATPase 3 |
| chr4_-_139084289 | 0.11 |
ENST00000510408.5
ENST00000379549.7 ENST00000358635.7 |
ELF2
|
E74 like ETS transcription factor 2 |
| chr11_-_125111708 | 0.11 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr1_+_23959797 | 0.11 |
ENST00000374468.1
ENST00000334351.8 |
PNRC2
|
proline rich nuclear receptor coactivator 2 |
| chr4_-_151227881 | 0.10 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr1_-_224434750 | 0.10 |
ENST00000414423.9
ENST00000678917.1 ENST00000678555.1 ENST00000678307.1 |
WDR26
|
WD repeat domain 26 |
| chr19_+_49850687 | 0.10 |
ENST00000594151.5
ENST00000600603.5 ENST00000601638.5 |
PTOV1
|
PTOV1 extended AT-hook containing adaptor protein |
| chr2_+_219245455 | 0.10 |
ENST00000409638.7
ENST00000396738.7 ENST00000409516.7 |
STK16
|
serine/threonine kinase 16 |
| chr11_+_65027402 | 0.10 |
ENST00000377244.8
ENST00000534637.5 ENST00000524831.5 |
SNX15
|
sorting nexin 15 |
| chr1_-_150697128 | 0.10 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr17_-_58531941 | 0.10 |
ENST00000581607.1
ENST00000317256.10 ENST00000426861.5 ENST00000580809.5 ENST00000577729.5 ENST00000583291.1 |
SEPTIN4
|
septin 4 |
| chr19_+_16067526 | 0.09 |
ENST00000646974.2
|
TPM4
|
tropomyosin 4 |
| chr17_-_59151794 | 0.09 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_+_46354072 | 0.09 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chr1_+_236394268 | 0.09 |
ENST00000334232.9
|
EDARADD
|
EDAR associated death domain |
| chr1_+_151721508 | 0.09 |
ENST00000479191.2
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit domain containing 1 |
| chr7_+_134891400 | 0.09 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr3_-_71493500 | 0.08 |
ENST00000648380.1
ENST00000650295.1 |
FOXP1
|
forkhead box P1 |
| chr1_-_92792396 | 0.08 |
ENST00000370331.5
ENST00000540033.2 |
EVI5
|
ecotropic viral integration site 5 |
| chr19_-_18543556 | 0.08 |
ENST00000544835.7
ENST00000608443.6 ENST00000597960.7 |
FKBP8
|
FKBP prolyl isomerase 8 |
| chr13_-_29850605 | 0.08 |
ENST00000380680.5
|
UBL3
|
ubiquitin like 3 |
| chr5_-_69369257 | 0.08 |
ENST00000509462.5
|
TAF9
|
TATA-box binding protein associated factor 9 |
| chr10_-_114144599 | 0.08 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chrX_-_116462965 | 0.08 |
ENST00000371894.5
|
CT83
|
cancer/testis antigen 83 |
| chr4_+_25914275 | 0.08 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr14_-_75069478 | 0.08 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr16_-_88703611 | 0.08 |
ENST00000541206.6
|
RNF166
|
ring finger protein 166 |
| chr3_+_38305936 | 0.08 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22 member 14 |
| chr1_-_110391041 | 0.08 |
ENST00000369781.8
ENST00000437429.6 ENST00000541986.5 |
SLC16A4
|
solute carrier family 16 member 4 |
| chr14_+_34993240 | 0.08 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr8_+_74320613 | 0.08 |
ENST00000675821.1
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr17_-_28576882 | 0.08 |
ENST00000395319.7
ENST00000581807.5 ENST00000226253.9 ENST00000584086.5 ENST00000395321.6 |
ALDOC
|
aldolase, fructose-bisphosphate C |
| chr22_-_36239517 | 0.07 |
ENST00000358502.10
ENST00000451256.6 |
APOL2
|
apolipoprotein L2 |
| chr14_-_70416994 | 0.07 |
ENST00000621525.4
ENST00000256366.6 |
SYNJ2BP-COX16
SYNJ2BP
|
SYNJ2BP-COX16 readthrough synaptojanin 2 binding protein |
| chr2_-_2326161 | 0.07 |
ENST00000649810.1
ENST00000648318.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr6_+_31971831 | 0.07 |
ENST00000375331.7
ENST00000375333.3 |
STK19
|
serine/threonine kinase 19 |
| chr7_+_143954844 | 0.07 |
ENST00000641412.1
|
OR2F1
|
olfactory receptor family 2 subfamily F member 1 |
| chr4_+_25914171 | 0.07 |
ENST00000506197.2
|
SMIM20
|
small integral membrane protein 20 |
| chr11_+_64924673 | 0.07 |
ENST00000164133.7
ENST00000532850.1 |
PPP2R5B
|
protein phosphatase 2 regulatory subunit B'beta |
| chrX_+_22272913 | 0.07 |
ENST00000323684.4
|
CBLL2
|
Cbl proto-oncogene like 2 |
| chr12_+_93677556 | 0.07 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr9_+_69123009 | 0.07 |
ENST00000647986.1
|
TJP2
|
tight junction protein 2 |
| chr1_-_152036984 | 0.07 |
ENST00000271638.3
|
S100A11
|
S100 calcium binding protein A11 |
| chr2_-_55296361 | 0.07 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr7_+_17299234 | 0.07 |
ENST00000637807.1
|
ENSG00000283321.1
|
novel protein |
| chr14_-_23286082 | 0.07 |
ENST00000357460.7
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr17_+_44004604 | 0.06 |
ENST00000293404.8
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr19_-_8149586 | 0.06 |
ENST00000600128.6
|
FBN3
|
fibrillin 3 |
| chr17_-_10469558 | 0.06 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr15_-_65115185 | 0.06 |
ENST00000559089.6
|
UBAP1L
|
ubiquitin associated protein 1 like |
| chr8_-_94208548 | 0.06 |
ENST00000027335.8
ENST00000441892.6 ENST00000521491.1 |
CDH17
|
cadherin 17 |
| chr19_-_58573555 | 0.06 |
ENST00000599369.5
|
MZF1
|
myeloid zinc finger 1 |
| chr9_-_4666495 | 0.06 |
ENST00000475086.5
|
SPATA6L
|
spermatogenesis associated 6 like |
| chr7_+_133253064 | 0.06 |
ENST00000393161.6
ENST00000253861.5 |
EXOC4
|
exocyst complex component 4 |
| chr19_+_47332167 | 0.06 |
ENST00000595464.3
|
C5AR2
|
complement component 5a receptor 2 |
| chr9_-_120793377 | 0.06 |
ENST00000684001.1
ENST00000684405.1 ENST00000608872.6 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr5_+_177134159 | 0.05 |
ENST00000511258.5
ENST00000347982.8 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr20_-_2664186 | 0.05 |
ENST00000474315.5
ENST00000380843.9 ENST00000380851.9 ENST00000613370.1 |
IDH3B
|
isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit beta |
| chr6_-_31972290 | 0.05 |
ENST00000375349.7
|
DXO
|
decapping exoribonuclease |
| chr9_-_127950716 | 0.05 |
ENST00000373084.8
|
FAM102A
|
family with sequence similarity 102 member A |
| chr19_-_58573280 | 0.05 |
ENST00000594234.5
ENST00000596039.1 ENST00000215057.7 |
MZF1
|
myeloid zinc finger 1 |
| chr14_-_50532518 | 0.05 |
ENST00000682126.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_+_99408958 | 0.05 |
ENST00000222969.10
|
BUD31
|
BUD31 homolog |
| chr11_-_31806886 | 0.05 |
ENST00000640963.1
|
PAX6
|
paired box 6 |
| chr16_-_67247460 | 0.05 |
ENST00000258201.9
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr1_+_116111395 | 0.05 |
ENST00000684484.1
ENST00000369500.4 |
MAB21L3
|
mab-21 like 3 |
| chr1_+_206470463 | 0.05 |
ENST00000581977.7
ENST00000578328.6 ENST00000584998.5 ENST00000579827.6 |
IKBKE
|
inhibitor of nuclear factor kappa B kinase subunit epsilon |
| chr14_+_67240713 | 0.05 |
ENST00000677382.1
|
MPP5
|
membrane palmitoylated protein 5 |
| chr11_-_41459592 | 0.04 |
ENST00000528697.6
ENST00000530763.5 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr1_-_31919093 | 0.04 |
ENST00000602683.5
ENST00000470404.5 |
PTP4A2
|
protein tyrosine phosphatase 4A2 |
| chr4_+_118034480 | 0.04 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr6_-_31972123 | 0.04 |
ENST00000337523.10
|
DXO
|
decapping exoribonuclease |
| chrX_+_71366222 | 0.04 |
ENST00000683202.1
ENST00000373790.9 |
TAF1
|
TATA-box binding protein associated factor 1 |
| chr5_+_139293728 | 0.04 |
ENST00000510056.5
ENST00000511249.5 ENST00000394805.8 ENST00000503811.5 ENST00000618441.5 |
MATR3
|
matrin 3 |
| chrX_+_71366290 | 0.04 |
ENST00000683782.1
ENST00000423759.6 |
TAF1
|
TATA-box binding protein associated factor 1 |
| chr20_-_37158904 | 0.04 |
ENST00000417458.5
|
MROH8
|
maestro heat like repeat family member 8 |
| chr11_-_31807617 | 0.04 |
ENST00000639920.1
ENST00000640460.1 |
PAX6
|
paired box 6 |
| chr6_-_27838362 | 0.04 |
ENST00000618958.2
|
H2AC15
|
H2A clustered histone 15 |
| chr1_-_150010675 | 0.04 |
ENST00000417191.2
ENST00000581312.6 |
OTUD7B
|
OTU deubiquitinase 7B |
| chr17_+_44847905 | 0.04 |
ENST00000587021.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr6_-_34671918 | 0.03 |
ENST00000374021.1
|
ILRUN
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr17_-_28812224 | 0.03 |
ENST00000582059.6
|
FAM222B
|
family with sequence similarity 222 member B |
| chr14_+_44962177 | 0.03 |
ENST00000361462.7
ENST00000361577.7 |
TOGARAM1
|
TOG array regulator of axonemal microtubules 1 |
| chr14_-_44961889 | 0.03 |
ENST00000579157.1
ENST00000396128.9 ENST00000556500.1 |
KLHL28
|
kelch like family member 28 |
| chr17_-_31314066 | 0.03 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr5_-_138331770 | 0.03 |
ENST00000415130.6
ENST00000323760.11 ENST00000503022.5 |
CDC25C
|
cell division cycle 25C |
| chr10_-_51699559 | 0.03 |
ENST00000331173.6
|
CSTF2T
|
cleavage stimulation factor subunit 2 tau variant |
| chr20_-_50153637 | 0.03 |
ENST00000341698.2
ENST00000371652.9 ENST00000371650.9 |
PEDS1-UBE2V1
PEDS1
|
PEDS1-UBE2V1 readthrough plasmanylethanolamine desaturase 1 |
| chr4_+_186266183 | 0.03 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr3_-_121749704 | 0.03 |
ENST00000393667.7
ENST00000340645.9 ENST00000614479.4 |
GOLGB1
|
golgin B1 |
| chr11_-_125111579 | 0.03 |
ENST00000532156.5
ENST00000532407.5 ENST00000279968.8 ENST00000527766.5 ENST00000529583.5 ENST00000524373.5 ENST00000527271.5 ENST00000526175.5 ENST00000529609.5 ENST00000682305.1 ENST00000533273.1 |
TMEM218
|
transmembrane protein 218 |
| chr12_+_54280842 | 0.03 |
ENST00000678077.1
ENST00000548688.5 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr1_-_159076742 | 0.03 |
ENST00000368130.9
|
AIM2
|
absent in melanoma 2 |
| chrX_+_136648138 | 0.03 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr17_+_28473635 | 0.03 |
ENST00000314669.10
ENST00000545060.2 |
SLC13A2
|
solute carrier family 13 member 2 |
| chr6_-_27146841 | 0.03 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr6_-_73354255 | 0.03 |
ENST00000370370.4
|
DPPA5
|
developmental pluripotency associated 5 |
| chr11_+_117327829 | 0.03 |
ENST00000533153.5
ENST00000278935.8 ENST00000525416.5 |
CEP164
|
centrosomal protein 164 |
| chr5_-_69369988 | 0.03 |
ENST00000380818.7
ENST00000328663.8 |
AK6
TAF9
|
adenylate kinase 6 TATA-box binding protein associated factor 9 |
| chrX_+_71283577 | 0.02 |
ENST00000420903.6
ENST00000373856.8 ENST00000678437.1 ENST00000678830.1 ENST00000677879.1 ENST00000373841.5 ENST00000276079.13 ENST00000676797.1 ENST00000678660.1 ENST00000678231.1 ENST00000677612.1 ENST00000413858.5 ENST00000450092.6 |
NONO
|
non-POU domain containing octamer binding |
| chr19_-_7489003 | 0.02 |
ENST00000221480.6
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr14_-_89954518 | 0.02 |
ENST00000556005.1
ENST00000555872.5 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr7_-_120857124 | 0.02 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr20_-_56005466 | 0.02 |
ENST00000064571.3
|
CBLN4
|
cerebellin 4 precursor |
| chr9_+_33290493 | 0.02 |
ENST00000379540.8
ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr3_+_33114007 | 0.02 |
ENST00000320954.11
|
CRTAP
|
cartilage associated protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMBOX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.3 | 2.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.2 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 0.7 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:2000439 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0070761 | PCAF complex(GO:0000125) pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 2.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |