Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
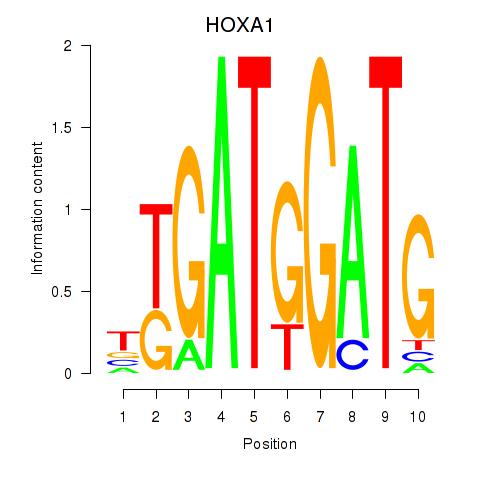
Results for HOXA1
Z-value: 0.81
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.10 | homeobox A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA1 | hg38_v1_chr7_-_27095972_27096039 | 0.84 | 9.8e-03 | Click! |
Activity profile of HOXA1 motif
Sorted Z-values of HOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10587219 | 3.91 |
ENST00000591240.5
ENST00000589684.5 ENST00000591676.1 ENST00000250244.11 ENST00000590923.5 |
AP1M2
|
adaptor related protein complex 1 subunit mu 2 |
| chr16_+_71626149 | 1.60 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr8_+_94641074 | 1.57 |
ENST00000423620.6
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr1_+_2073986 | 1.21 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr16_+_71626175 | 1.17 |
ENST00000268485.8
ENST00000565261.1 ENST00000299952.4 |
MARVELD3
|
MARVEL domain containing 3 |
| chr5_+_167754918 | 1.15 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chr12_-_95116967 | 1.03 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr20_+_59628609 | 0.90 |
ENST00000541461.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr1_+_183186238 | 0.87 |
ENST00000493293.5
ENST00000264144.5 |
LAMC2
|
laminin subunit gamma 2 |
| chr1_-_201399525 | 0.85 |
ENST00000367313.4
|
LAD1
|
ladinin 1 |
| chr11_+_18266254 | 0.81 |
ENST00000532858.5
ENST00000649195.1 ENST00000356524.9 ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr6_+_130366281 | 0.74 |
ENST00000617887.4
|
TMEM200A
|
transmembrane protein 200A |
| chr7_-_22193824 | 0.60 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr16_+_83968244 | 0.55 |
ENST00000305202.9
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr6_+_89562308 | 0.53 |
ENST00000522441.5
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr19_+_53867874 | 0.53 |
ENST00000448420.5
ENST00000439000.5 ENST00000391771.1 ENST00000391770.9 |
MYADM
|
myeloid associated differentiation marker |
| chr12_-_84912705 | 0.49 |
ENST00000679933.1
ENST00000680260.1 ENST00000551010.2 ENST00000679453.1 ENST00000681281.1 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr7_-_22193728 | 0.46 |
ENST00000620335.4
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr8_-_90082871 | 0.45 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr5_+_140834230 | 0.44 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr10_-_103855406 | 0.44 |
ENST00000355946.6
ENST00000369774.8 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr12_-_52517929 | 0.42 |
ENST00000548409.5
|
KRT5
|
keratin 5 |
| chr2_-_75560893 | 0.42 |
ENST00000410113.5
ENST00000393913.8 |
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chr2_-_70835808 | 0.41 |
ENST00000410009.5
|
CD207
|
CD207 molecule |
| chr15_-_52295792 | 0.41 |
ENST00000261839.12
|
MYO5C
|
myosin VC |
| chr12_-_84912783 | 0.40 |
ENST00000680892.1
ENST00000266682.10 ENST00000680714.1 ENST00000552192.5 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr3_-_112845950 | 0.39 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr20_-_23637933 | 0.38 |
ENST00000398411.5
|
CST3
|
cystatin C |
| chr6_-_13487593 | 0.38 |
ENST00000379287.4
ENST00000603223.1 |
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr9_-_5304713 | 0.38 |
ENST00000381627.4
|
RLN2
|
relaxin 2 |
| chr11_-_72642450 | 0.38 |
ENST00000444035.6
ENST00000544570.5 |
PDE2A
|
phosphodiesterase 2A |
| chr11_-_72642407 | 0.37 |
ENST00000376450.7
|
PDE2A
|
phosphodiesterase 2A |
| chr1_+_205504592 | 0.37 |
ENST00000506784.5
ENST00000360066.6 |
CDK18
|
cyclin dependent kinase 18 |
| chrX_+_135309480 | 0.36 |
ENST00000635820.1
|
ETDC
|
embryonic testis differentiation homolog C |
| chr11_-_5441514 | 0.36 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1 |
| chr19_-_38256513 | 0.36 |
ENST00000347262.8
ENST00000591585.1 |
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr7_+_20330893 | 0.35 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr15_-_63381835 | 0.33 |
ENST00000344366.7
ENST00000178638.8 ENST00000422263.2 |
CA12
|
carbonic anhydrase 12 |
| chr1_+_44674688 | 0.33 |
ENST00000418644.5
ENST00000458657.6 ENST00000535358.6 ENST00000441519.5 ENST00000445071.5 |
ARMH1
|
armadillo like helical domain containing 1 |
| chr1_-_24143112 | 0.33 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr1_+_18630839 | 0.32 |
ENST00000420770.7
|
PAX7
|
paired box 7 |
| chr16_+_2988256 | 0.32 |
ENST00000573315.2
|
LINC00514
|
long intergenic non-protein coding RNA 514 |
| chr11_+_2461432 | 0.32 |
ENST00000335475.6
|
KCNQ1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr6_-_107115493 | 0.30 |
ENST00000369042.6
|
BEND3
|
BEN domain containing 3 |
| chr1_+_202203721 | 0.28 |
ENST00000255432.11
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6 |
| chr6_-_47042260 | 0.28 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr3_-_129893551 | 0.24 |
ENST00000505616.5
ENST00000426664.6 ENST00000648771.1 ENST00000393238.8 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr8_+_97775775 | 0.24 |
ENST00000521545.7
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr9_+_121567057 | 0.24 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr2_+_1484663 | 0.23 |
ENST00000446278.5
ENST00000469607.3 |
TPO
|
thyroid peroxidase |
| chr8_+_97775829 | 0.23 |
ENST00000517924.5
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr18_-_76495191 | 0.23 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chrX_+_135520616 | 0.22 |
ENST00000370752.4
ENST00000639893.2 |
INTS6L
|
integrator complex subunit 6 like |
| chr3_-_179243284 | 0.22 |
ENST00000486944.2
|
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr1_-_206921867 | 0.21 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr1_+_156154371 | 0.20 |
ENST00000368282.1
|
SEMA4A
|
semaphorin 4A |
| chr8_+_2045058 | 0.20 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr8_+_142449430 | 0.20 |
ENST00000643448.1
ENST00000517894.5 |
ADGRB1
|
adhesion G protein-coupled receptor B1 |
| chr11_-_69819410 | 0.20 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr21_+_31873010 | 0.19 |
ENST00000270112.7
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr8_+_85187650 | 0.19 |
ENST00000517476.5
ENST00000521429.5 |
E2F5
|
E2F transcription factor 5 |
| chr6_+_15248855 | 0.19 |
ENST00000397311.4
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr3_+_133038366 | 0.19 |
ENST00000321871.11
ENST00000393130.7 ENST00000514894.5 ENST00000512662.5 |
TMEM108
|
transmembrane protein 108 |
| chr5_+_36151989 | 0.19 |
ENST00000274254.9
|
SKP2
|
S-phase kinase associated protein 2 |
| chr1_-_206921987 | 0.18 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr17_-_15598618 | 0.18 |
ENST00000583965.5
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr1_-_112956063 | 0.18 |
ENST00000538576.5
ENST00000369626.8 ENST00000458229.6 |
SLC16A1
|
solute carrier family 16 member 1 |
| chr2_-_60550900 | 0.17 |
ENST00000643222.1
ENST00000643459.1 ENST00000489516.7 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr14_+_24130659 | 0.17 |
ENST00000267426.6
|
FITM1
|
fat storage inducing transmembrane protein 1 |
| chr15_+_47717344 | 0.17 |
ENST00000558816.5
ENST00000536845.7 |
SEMA6D
|
semaphorin 6D |
| chr15_+_66453418 | 0.17 |
ENST00000566326.1
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr16_+_75222609 | 0.17 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr8_-_118111806 | 0.16 |
ENST00000378204.7
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr4_-_71784046 | 0.16 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr3_-_161372821 | 0.16 |
ENST00000617024.1
ENST00000359175.8 |
SPTSSB
|
serine palmitoyltransferase small subunit B |
| chr4_+_70334963 | 0.15 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr16_+_28974764 | 0.15 |
ENST00000565975.5
ENST00000311008.16 ENST00000323081.12 ENST00000334536.12 |
SPNS1
|
sphingolipid transporter 1 (putative) |
| chr15_-_22185402 | 0.15 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr16_+_30023198 | 0.15 |
ENST00000681219.1
ENST00000300575.6 |
C16orf92
|
chromosome 16 open reading frame 92 |
| chr7_+_98106852 | 0.14 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2 |
| chrX_+_83508284 | 0.14 |
ENST00000644024.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr19_-_51531790 | 0.14 |
ENST00000359982.8
ENST00000436458.5 ENST00000391797.3 ENST00000343300.8 |
SIGLEC6
|
sialic acid binding Ig like lectin 6 |
| chr12_+_124993633 | 0.14 |
ENST00000341446.9
ENST00000671775.2 |
BRI3BP
|
BRI3 binding protein |
| chr7_+_139829242 | 0.14 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr17_-_46818680 | 0.14 |
ENST00000225512.6
|
WNT3
|
Wnt family member 3 |
| chr2_+_85429448 | 0.14 |
ENST00000651736.1
|
SH2D6
|
SH2 domain containing 6 |
| chr12_+_75334655 | 0.14 |
ENST00000378695.9
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr12_+_75334675 | 0.14 |
ENST00000312442.2
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr9_+_122519141 | 0.14 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor family 1 subfamily J member 4 |
| chr4_-_86101922 | 0.14 |
ENST00000472236.5
ENST00000641881.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_-_15898057 | 0.14 |
ENST00000011989.11
ENST00000221700.11 |
CYP4F2
|
cytochrome P450 family 4 subfamily F member 2 |
| chr12_-_10810168 | 0.14 |
ENST00000240691.4
|
TAS2R9
|
taste 2 receptor member 9 |
| chr13_-_19503277 | 0.14 |
ENST00000382978.5
ENST00000400230.6 ENST00000255310.10 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr17_-_3595831 | 0.14 |
ENST00000399759.7
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chrX_+_129779930 | 0.13 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr19_-_11339573 | 0.13 |
ENST00000222120.8
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr5_+_36152077 | 0.13 |
ENST00000546211.6
ENST00000620197.5 ENST00000678270.1 ENST00000679015.1 ENST00000678580.1 ENST00000274255.11 ENST00000508514.5 |
SKP2
|
S-phase kinase associated protein 2 |
| chr19_+_15049469 | 0.13 |
ENST00000427043.4
|
CASP14
|
caspase 14 |
| chr15_-_74208969 | 0.13 |
ENST00000423167.6
ENST00000432245.6 |
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr10_+_97589715 | 0.12 |
ENST00000370640.5
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr17_-_40665121 | 0.12 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr4_-_38804783 | 0.12 |
ENST00000308979.7
ENST00000505940.1 ENST00000515861.5 |
TLR1
|
toll like receptor 1 |
| chr13_-_20161038 | 0.12 |
ENST00000241125.4
|
GJA3
|
gap junction protein alpha 3 |
| chr2_+_72887368 | 0.12 |
ENST00000234454.6
|
SPR
|
sepiapterin reductase |
| chr5_-_784691 | 0.12 |
ENST00000508859.8
ENST00000652055.1 ENST00000651083.1 |
ZDHHC11B
|
zinc finger DHHC-type containing 11B |
| chr19_+_5681140 | 0.12 |
ENST00000579559.1
ENST00000577222.5 |
HSD11B1L
RPL36
|
hydroxysteroid 11-beta dehydrogenase 1 like ribosomal protein L36 |
| chr3_+_48223479 | 0.12 |
ENST00000652295.2
|
CAMP
|
cathelicidin antimicrobial peptide |
| chr9_-_6645712 | 0.12 |
ENST00000321612.8
|
GLDC
|
glycine decarboxylase |
| chr16_+_23835946 | 0.12 |
ENST00000321728.12
ENST00000643927.1 |
PRKCB
|
protein kinase C beta |
| chr6_+_29170907 | 0.12 |
ENST00000641417.1
|
OR2J2
|
olfactory receptor family 2 subfamily J member 2 |
| chr16_+_29663219 | 0.11 |
ENST00000436527.5
ENST00000360121.4 ENST00000652691.1 ENST00000449759.2 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr7_-_117323041 | 0.11 |
ENST00000491214.1
ENST00000265441.8 |
WNT2
|
Wnt family member 2 |
| chr7_+_80135694 | 0.11 |
ENST00000457358.7
|
GNAI1
|
G protein subunit alpha i1 |
| chr17_-_44199834 | 0.11 |
ENST00000587097.6
|
ATXN7L3
|
ataxin 7 like 3 |
| chr3_+_31532901 | 0.11 |
ENST00000295770.4
|
STT3B
|
STT3 oligosaccharyltransferase complex catalytic subunit B |
| chr11_-_18791768 | 0.11 |
ENST00000358540.7
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5 |
| chr20_+_2692736 | 0.11 |
ENST00000380648.9
ENST00000497450.5 |
EBF4
|
EBF family member 4 |
| chr1_-_44141631 | 0.11 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr2_-_49154507 | 0.11 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor |
| chr11_+_4704782 | 0.10 |
ENST00000380390.6
|
MMP26
|
matrix metallopeptidase 26 |
| chr1_+_201283452 | 0.10 |
ENST00000263946.7
ENST00000367324.8 |
PKP1
|
plakophilin 1 |
| chr9_+_110668779 | 0.10 |
ENST00000416899.7
ENST00000374448.9 |
MUSK
|
muscle associated receptor tyrosine kinase |
| chr13_-_35476682 | 0.10 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chr11_+_68460712 | 0.10 |
ENST00000528635.5
ENST00000533127.5 ENST00000529907.5 ENST00000529344.5 ENST00000534534.5 ENST00000524845.5 ENST00000393800.7 ENST00000265637.8 ENST00000524904.5 ENST00000393801.7 ENST00000265636.9 ENST00000529710.5 |
PPP6R3
|
protein phosphatase 6 regulatory subunit 3 |
| chr2_-_49154433 | 0.10 |
ENST00000454032.5
ENST00000304421.8 |
FSHR
|
follicle stimulating hormone receptor |
| chr9_-_127916978 | 0.10 |
ENST00000361444.3
ENST00000335791.10 |
ST6GALNAC4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr10_+_7703340 | 0.10 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_-_5227063 | 0.10 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr3_+_4493340 | 0.10 |
ENST00000357086.10
ENST00000354582.12 ENST00000649015.2 ENST00000467056.6 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr3_+_150546671 | 0.10 |
ENST00000487799.5
|
EIF2A
|
eukaryotic translation initiation factor 2A |
| chrX_+_8465426 | 0.09 |
ENST00000381029.4
|
VCX3B
|
variable charge X-linked 3B |
| chr7_-_101165114 | 0.09 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr18_-_3874247 | 0.09 |
ENST00000581699.5
|
DLGAP1
|
DLG associated protein 1 |
| chr16_+_28974813 | 0.09 |
ENST00000352260.11
|
SPNS1
|
sphingolipid transporter 1 (putative) |
| chr1_-_44788168 | 0.09 |
ENST00000372207.4
|
BEST4
|
bestrophin 4 |
| chr4_+_113049616 | 0.09 |
ENST00000504454.5
ENST00000357077.9 ENST00000394537.7 ENST00000672779.1 ENST00000264366.10 |
ANK2
|
ankyrin 2 |
| chr5_-_137735997 | 0.09 |
ENST00000505853.1
|
KLHL3
|
kelch like family member 3 |
| chr3_+_150546765 | 0.09 |
ENST00000406576.7
ENST00000460851.6 ENST00000482093.5 ENST00000273435.9 |
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr20_+_37383648 | 0.09 |
ENST00000373567.6
|
SRC
|
SRC proto-oncogene, non-receptor tyrosine kinase |
| chr2_-_27356975 | 0.09 |
ENST00000423998.1
ENST00000264720.7 |
GTF3C2
|
general transcription factor IIIC subunit 2 |
| chr12_+_57216779 | 0.08 |
ENST00000349394.6
|
NXPH4
|
neurexophilin 4 |
| chr3_+_4493442 | 0.08 |
ENST00000456211.8
ENST00000443694.5 ENST00000648266.1 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chrM_+_4467 | 0.08 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr3_-_114199407 | 0.08 |
ENST00000460779.5
|
DRD3
|
dopamine receptor D3 |
| chrX_-_73214793 | 0.08 |
ENST00000373517.4
|
NAP1L2
|
nucleosome assembly protein 1 like 2 |
| chr18_-_3874270 | 0.08 |
ENST00000400149.7
ENST00000400155.5 ENST00000400150.7 |
DLGAP1
|
DLG associated protein 1 |
| chr11_+_66480007 | 0.08 |
ENST00000531863.5
ENST00000532677.5 |
DPP3
|
dipeptidyl peptidase 3 |
| chr9_+_100429511 | 0.08 |
ENST00000613183.1
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr8_-_113436883 | 0.08 |
ENST00000455883.2
ENST00000297405.10 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chrX_-_10677720 | 0.08 |
ENST00000453318.6
|
MID1
|
midline 1 |
| chr13_-_77919390 | 0.08 |
ENST00000475537.2
ENST00000646605.1 |
EDNRB
|
endothelin receptor type B |
| chr9_+_110668854 | 0.08 |
ENST00000189978.10
ENST00000374440.7 |
MUSK
|
muscle associated receptor tyrosine kinase |
| chr7_-_101165558 | 0.08 |
ENST00000611537.1
ENST00000249330.3 |
VGF
|
VGF nerve growth factor inducible |
| chr3_+_179347686 | 0.08 |
ENST00000471841.6
|
MFN1
|
mitofusin 1 |
| chr10_+_7703300 | 0.07 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr7_+_154305105 | 0.07 |
ENST00000332007.7
|
DPP6
|
dipeptidyl peptidase like 6 |
| chr11_+_55635113 | 0.07 |
ENST00000641760.1
|
OR4P4
|
olfactory receptor family 4 subfamily P member 4 |
| chrX_+_37780049 | 0.07 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chrX_+_15507302 | 0.07 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr1_-_217631034 | 0.07 |
ENST00000366934.3
ENST00000366935.8 |
GPATCH2
|
G-patch domain containing 2 |
| chr14_-_68979076 | 0.07 |
ENST00000538545.6
ENST00000684639.1 |
ACTN1
|
actinin alpha 1 |
| chr16_-_8936633 | 0.07 |
ENST00000381886.8
|
USP7
|
ubiquitin specific peptidase 7 |
| chr11_+_26188836 | 0.07 |
ENST00000672621.1
|
ANO3
|
anoctamin 3 |
| chr6_+_42155399 | 0.07 |
ENST00000623004.2
ENST00000372963.4 ENST00000654459.1 |
GUCA1ANB
GUCA1A
|
GUCA1A neighbor guanylate cyclase activator 1A |
| chrX_+_116170742 | 0.07 |
ENST00000371906.5
ENST00000681852.1 |
AGTR2
|
angiotensin II receptor type 2 |
| chr11_-_61878582 | 0.07 |
ENST00000527379.5
|
FADS3
|
fatty acid desaturase 3 |
| chr12_-_101210232 | 0.07 |
ENST00000536262.3
|
SLC5A8
|
solute carrier family 5 member 8 |
| chr1_+_43300971 | 0.07 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr17_+_62370218 | 0.07 |
ENST00000450662.7
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr12_+_8123899 | 0.07 |
ENST00000641376.1
|
CLEC4A
|
C-type lectin domain family 4 member A |
| chr14_-_68979274 | 0.07 |
ENST00000394419.9
|
ACTN1
|
actinin alpha 1 |
| chr5_-_137736066 | 0.07 |
ENST00000309755.9
|
KLHL3
|
kelch like family member 3 |
| chr1_-_12848720 | 0.06 |
ENST00000317869.7
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C like 1 |
| chr15_-_43220989 | 0.06 |
ENST00000540029.5
ENST00000441366.7 ENST00000648595.1 |
EPB42
|
erythrocyte membrane protein band 4.2 |
| chr14_-_68979251 | 0.06 |
ENST00000438964.6
ENST00000679147.1 |
ACTN1
|
actinin alpha 1 |
| chr20_+_45408276 | 0.06 |
ENST00000372710.5
ENST00000443296.1 |
DBNDD2
|
dysbindin domain containing 2 |
| chr12_-_21774688 | 0.06 |
ENST00000240662.3
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr5_+_64505981 | 0.06 |
ENST00000334025.3
|
RGS7BP
|
regulator of G protein signaling 7 binding protein |
| chr12_-_86256267 | 0.06 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr15_+_73443149 | 0.06 |
ENST00000560581.1
ENST00000331090.11 |
REC114
|
REC114 meiotic recombination protein |
| chr8_+_42338477 | 0.06 |
ENST00000518925.5
ENST00000265421.9 |
POLB
|
DNA polymerase beta |
| chr3_+_44874606 | 0.06 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr14_-_68979314 | 0.06 |
ENST00000684713.1
ENST00000683198.1 ENST00000684598.1 ENST00000682331.1 ENST00000682291.1 ENST00000683342.1 |
ACTN1
|
actinin alpha 1 |
| chr11_-_18791563 | 0.06 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5 |
| chr11_+_125904467 | 0.06 |
ENST00000263576.11
ENST00000530414.5 ENST00000530129.6 |
DDX25
|
DEAD-box helicase 25 |
| chr6_-_31592952 | 0.06 |
ENST00000376073.8
ENST00000376072.7 |
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr8_+_40153475 | 0.06 |
ENST00000315792.5
|
TCIM
|
transcriptional and immune response regulator |
| chr13_+_29428603 | 0.06 |
ENST00000380808.6
|
MTUS2
|
microtubule associated scaffold protein 2 |
| chr16_-_48610150 | 0.06 |
ENST00000262384.4
|
N4BP1
|
NEDD4 binding protein 1 |
| chr9_-_35563867 | 0.06 |
ENST00000399742.7
ENST00000619051.4 |
FAM166B
|
family with sequence similarity 166 member B |
| chr13_+_113209597 | 0.06 |
ENST00000488558.2
ENST00000375440.9 |
CUL4A
|
cullin 4A |
| chr12_+_63779894 | 0.06 |
ENST00000261234.11
|
RXYLT1
|
ribitol xylosyltransferase 1 |
| chr3_+_111542178 | 0.06 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr1_+_36156096 | 0.05 |
ENST00000474796.2
ENST00000373150.8 ENST00000373151.6 |
MAP7D1
|
MAP7 domain containing 1 |
| chr6_-_41154326 | 0.05 |
ENST00000426005.6
ENST00000437044.2 ENST00000373127.8 |
TREML1
|
triggering receptor expressed on myeloid cells like 1 |
| chr17_+_80178499 | 0.05 |
ENST00000570421.5
ENST00000344227.6 |
CARD14
|
caspase recruitment domain family member 14 |
| chr2_-_224569782 | 0.05 |
ENST00000409096.5
|
CUL3
|
cullin 3 |
| chr1_-_110519175 | 0.05 |
ENST00000369771.4
|
KCNA10
|
potassium voltage-gated channel subfamily A member 10 |
| chr17_+_42780592 | 0.05 |
ENST00000246914.10
|
WNK4
|
WNK lysine deficient protein kinase 4 |
| chr2_-_50347789 | 0.05 |
ENST00000628364.2
|
NRXN1
|
neurexin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 1.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.7 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.6 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 0.3 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.1 | 0.3 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 0.4 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.9 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 3.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.2 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.0 | 0.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.2 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.8 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0045553 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.9 | GO:0043256 | laminin complex(GO:0043256) |
| 0.1 | 3.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.1 | 0.9 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 3.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.7 | GO:0030911 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) TPR domain binding(GO:0030911) |
| 0.1 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 1.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |