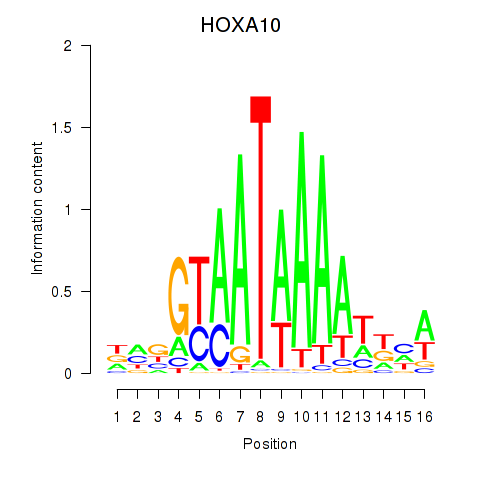
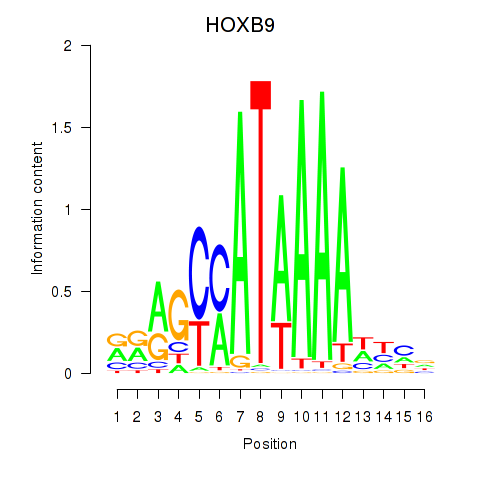
|
chr7_-_41703062
Show fit
|
0.73 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A
|
|
chr1_+_160400543
Show fit
|
0.50 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2
|
|
chr1_+_153031195
Show fit
|
0.49 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B
|
|
chr1_+_244352627
Show fit
|
0.43 |
ENST00000366537.5
ENST00000308105.5
|
C1orf100
|
chromosome 1 open reading frame 100
|
|
chr5_+_36608146
Show fit
|
0.41 |
ENST00000381918.4
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 member 3
|
|
chr17_-_41489907
Show fit
|
0.41 |
ENST00000328119.11
|
KRT36
|
keratin 36
|
|
chr2_-_51032151
Show fit
|
0.37 |
ENST00000628515.2
|
NRXN1
|
neurexin 1
|
|
chr7_+_129144691
Show fit
|
0.35 |
ENST00000486685.3
|
TSPAN33
|
tetraspanin 33
|
|
chr5_+_36596583
Show fit
|
0.35 |
ENST00000680318.1
|
SLC1A3
|
solute carrier family 1 member 3
|
|
chr5_+_140786136
Show fit
|
0.33 |
ENST00000378133.4
ENST00000504120.4
|
PCDHA1
|
protocadherin alpha 1
|
|
chr10_-_127892930
Show fit
|
0.29 |
ENST00000368671.4
|
CLRN3
|
clarin 3
|
|
chr5_+_140882116
Show fit
|
0.28 |
ENST00000289272.3
ENST00000409494.5
ENST00000617769.1
|
PCDHA13
|
protocadherin alpha 13
|
|
chr5_+_36606355
Show fit
|
0.27 |
ENST00000681909.1
ENST00000513903.5
ENST00000681795.1
ENST00000680125.1
ENST00000612708.5
ENST00000680232.1
ENST00000681776.1
ENST00000681926.1
ENST00000679958.1
ENST00000265113.9
ENST00000504121.5
ENST00000512374.1
ENST00000613445.5
ENST00000679983.1
|
SLC1A3
|
solute carrier family 1 member 3
|
|
chr6_+_132538290
Show fit
|
0.26 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9
|
|
chr11_+_111245725
Show fit
|
0.26 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53
|
|
chr4_-_71784046
Show fit
|
0.25 |
ENST00000513476.5
ENST00000273951.13
|
GC
|
GC vitamin D binding protein
|
|
chr1_+_13585453
Show fit
|
0.24 |
ENST00000487038.5
ENST00000475043.5
|
PDPN
|
podoplanin
|
|
chr7_-_143408848
Show fit
|
0.23 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1
|
|
chr8_-_80171496
Show fit
|
0.22 |
ENST00000379096.9
ENST00000518937.6
|
TPD52
|
tumor protein D52
|
|
chr10_+_84173793
Show fit
|
0.21 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr11_+_63838902
Show fit
|
0.20 |
ENST00000377810.8
|
MARK2
|
microtubule affinity regulating kinase 2
|
|
chrX_+_106693838
Show fit
|
0.19 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128
|
|
chr15_-_43879835
Show fit
|
0.19 |
ENST00000636859.1
|
FRMD5
|
FERM domain containing 5
|
|
chr4_+_95051671
Show fit
|
0.18 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr12_-_84892120
Show fit
|
0.18 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15
|
|
chr18_+_58196736
Show fit
|
0.18 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr12_+_69825221
Show fit
|
0.17 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like
|
|
chr6_+_29301701
Show fit
|
0.16 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1
|
|
chr11_+_10450289
Show fit
|
0.16 |
ENST00000444303.6
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr2_-_216013582
Show fit
|
0.16 |
ENST00000620139.4
|
MREG
|
melanoregulin
|
|
chr19_+_4402615
Show fit
|
0.15 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A
|
|
chr5_+_140786291
Show fit
|
0.15 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1
|
|
chr2_+_113127588
Show fit
|
0.15 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr1_-_186461089
Show fit
|
0.15 |
ENST00000391997.3
|
PDC
|
phosducin
|
|
chr12_-_18090185
Show fit
|
0.14 |
ENST00000229002.6
ENST00000538724.6
|
RERGL
|
RERG like
|
|
chr1_+_23019415
Show fit
|
0.14 |
ENST00000465864.2
ENST00000356634.7
ENST00000400181.9
|
KDM1A
|
lysine demethylase 1A
|
|
chr6_+_43770707
Show fit
|
0.13 |
ENST00000324450.11
ENST00000417285.7
ENST00000413642.8
ENST00000372055.9
ENST00000482630.7
ENST00000425836.7
ENST00000372064.9
ENST00000372077.8
ENST00000519767.5
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr2_-_68952880
Show fit
|
0.13 |
ENST00000481498.1
ENST00000328895.9
|
GKN2
|
gastrokine 2
|
|
chr6_+_43770202
Show fit
|
0.13 |
ENST00000372067.8
ENST00000672860.2
|
VEGFA
|
vascular endothelial growth factor A
|
|
chrX_-_134658450
Show fit
|
0.13 |
ENST00000359237.9
|
PLAC1
|
placenta enriched 1
|
|
chr12_+_1691011
Show fit
|
0.12 |
ENST00000357103.5
|
ADIPOR2
|
adiponectin receptor 2
|
|
chr6_+_121435595
Show fit
|
0.12 |
ENST00000649003.1
ENST00000282561.4
|
GJA1
|
gap junction protein alpha 1
|
|
chr2_+_205683109
Show fit
|
0.12 |
ENST00000357118.8
ENST00000272849.7
ENST00000412873.2
|
NRP2
|
neuropilin 2
|
|
chr12_+_69825273
Show fit
|
0.12 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like
|
|
chr20_+_10035029
Show fit
|
0.11 |
ENST00000378380.4
|
ANKEF1
|
ankyrin repeat and EF-hand domain containing 1
|
|
chr20_+_10034963
Show fit
|
0.11 |
ENST00000378392.6
|
ANKEF1
|
ankyrin repeat and EF-hand domain containing 1
|
|
chr8_-_21812320
Show fit
|
0.11 |
ENST00000517328.5
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr4_+_70397931
Show fit
|
0.11 |
ENST00000399575.7
|
OPRPN
|
opiorphin prepropeptide
|
|
chr18_-_47035621
Show fit
|
0.11 |
ENST00000332567.6
|
ELOA2
|
elongin A2
|
|
chr4_+_143433491
Show fit
|
0.11 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1
|
|
chr3_+_57890011
Show fit
|
0.11 |
ENST00000494088.6
ENST00000438794.5
|
SLMAP
|
sarcolemma associated protein
|
|
chr18_-_77127935
Show fit
|
0.11 |
ENST00000581878.5
|
MBP
|
myelin basic protein
|
|
chr21_-_30829755
Show fit
|
0.10 |
ENST00000621162.1
|
KRTAP7-1
|
keratin associated protein 7-1
|
|
chr12_+_20810698
Show fit
|
0.10 |
ENST00000540853.5
ENST00000381545.8
|
SLCO1B3
|
solute carrier organic anion transporter family member 1B3
|
|
chr8_+_106726115
Show fit
|
0.10 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1
|
|
chr15_-_83283449
Show fit
|
0.10 |
ENST00000569704.2
|
BNC1
|
basonuclin 1
|
|
chr3_+_125969152
Show fit
|
0.10 |
ENST00000251776.8
ENST00000504401.1
|
ROPN1B
|
rhophilin associated tail protein 1B
|
|
chr21_-_14210884
Show fit
|
0.10 |
ENST00000679868.1
ENST00000400211.3
ENST00000680801.1
ENST00000536861.6
ENST00000614229.5
|
LIPI
|
lipase I
|
|
chr10_+_94938649
Show fit
|
0.10 |
ENST00000461906.1
ENST00000260682.8
|
CYP2C9
|
cytochrome P450 family 2 subfamily C member 9
|
|
chr20_+_44355692
Show fit
|
0.10 |
ENST00000316673.8
ENST00000609795.5
ENST00000457232.5
ENST00000609262.5
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr1_+_67307360
Show fit
|
0.10 |
ENST00000262345.5
ENST00000544434.5
|
IL12RB2
|
interleukin 12 receptor subunit beta 2
|
|
chr19_+_3185911
Show fit
|
0.09 |
ENST00000246117.9
ENST00000588428.5
|
NCLN
|
nicalin
|
|
chr18_-_55322215
Show fit
|
0.09 |
ENST00000457482.7
|
TCF4
|
transcription factor 4
|
|
chr2_-_51032091
Show fit
|
0.09 |
ENST00000637511.1
ENST00000405472.7
ENST00000405581.3
ENST00000401669.7
|
NRXN1
|
neurexin 1
|
|
chr21_-_14210948
Show fit
|
0.09 |
ENST00000681601.1
|
LIPI
|
lipase I
|
|
chr5_+_122129533
Show fit
|
0.08 |
ENST00000296600.5
ENST00000504912.1
ENST00000505843.1
|
ZNF474
|
zinc finger protein 474
|
|
chr18_-_55321986
Show fit
|
0.08 |
ENST00000570287.6
|
TCF4
|
transcription factor 4
|
|
chr10_-_72088533
Show fit
|
0.08 |
ENST00000373109.7
|
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2
|
|
chr8_+_106726012
Show fit
|
0.08 |
ENST00000449762.6
ENST00000297447.10
|
OXR1
|
oxidation resistance 1
|
|
chr4_-_89838289
Show fit
|
0.08 |
ENST00000336904.7
|
SNCA
|
synuclein alpha
|
|
chr1_-_226739271
Show fit
|
0.08 |
ENST00000429204.6
ENST00000366784.1
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr2_+_27217361
Show fit
|
0.08 |
ENST00000264705.9
ENST00000403525.5
|
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
|
|
chr16_-_1884231
Show fit
|
0.07 |
ENST00000563416.3
ENST00000633813.1
ENST00000470044.5
|
LINC00254
MEIOB
|
long intergenic non-protein coding RNA 254
meiosis specific with OB-fold
|
|
chr12_+_20815672
Show fit
|
0.07 |
ENST00000261196.6
ENST00000381541.7
ENST00000540229.1
|
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3
SLCO1B3-SLCO1B7 readthrough
|
|
chr15_+_40594001
Show fit
|
0.07 |
ENST00000346991.9
ENST00000528975.5
|
KNL1
|
kinetochore scaffold 1
|
|
chr15_+_66386902
Show fit
|
0.07 |
ENST00000307102.10
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr17_-_31297231
Show fit
|
0.07 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chrX_+_66164340
Show fit
|
0.07 |
ENST00000441993.7
ENST00000419594.6
ENST00000425114.2
|
HEPH
|
hephaestin
|
|
chr9_+_131190119
Show fit
|
0.07 |
ENST00000483497.6
|
NUP214
|
nucleoporin 214
|
|
chr17_-_10549652
Show fit
|
0.07 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2
|
|
chr2_+_233760265
Show fit
|
0.07 |
ENST00000305208.10
ENST00000360418.4
|
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1
|
|
chr6_+_25279359
Show fit
|
0.07 |
ENST00000329474.7
|
CARMIL1
|
capping protein regulator and myosin 1 linker 1
|
|
chr17_-_10549612
Show fit
|
0.07 |
ENST00000532183.6
ENST00000397183.6
ENST00000420805.1
|
MYH2
|
myosin heavy chain 2
|
|
chr9_+_122370523
Show fit
|
0.07 |
ENST00000643810.1
ENST00000540753.6
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr8_-_103501890
Show fit
|
0.07 |
ENST00000649416.1
|
ENSG00000285982.1
|
novel protein
|
|
chr12_-_70754631
Show fit
|
0.07 |
ENST00000440835.6
ENST00000549308.5
ENST00000550661.1
ENST00000378778.5
|
PTPRR
|
protein tyrosine phosphatase receptor type R
|
|
chr5_-_16738341
Show fit
|
0.07 |
ENST00000515803.5
|
MYO10
|
myosin X
|
|
chr2_+_196713117
Show fit
|
0.07 |
ENST00000409270.5
|
CCDC150
|
coiled-coil domain containing 150
|
|
chrX_+_66164210
Show fit
|
0.06 |
ENST00000343002.7
ENST00000336279.9
|
HEPH
|
hephaestin
|
|
chr17_-_10549694
Show fit
|
0.06 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2
|
|
chr3_-_197184131
Show fit
|
0.06 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr7_+_80133830
Show fit
|
0.06 |
ENST00000648098.1
ENST00000648476.1
ENST00000648412.1
ENST00000648953.1
ENST00000648306.1
ENST00000648832.1
ENST00000648877.1
ENST00000442586.2
ENST00000649487.1
ENST00000649267.1
|
GNAI1
|
G protein subunit alpha i1
|
|
chr7_-_50565381
Show fit
|
0.06 |
ENST00000444124.7
|
DDC
|
dopa decarboxylase
|
|
chr8_+_2045037
Show fit
|
0.06 |
ENST00000262113.9
|
MYOM2
|
myomesin 2
|
|
chr8_+_2045058
Show fit
|
0.06 |
ENST00000523438.1
|
MYOM2
|
myomesin 2
|
|
chr6_+_131637296
Show fit
|
0.06 |
ENST00000358229.6
ENST00000357639.8
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr9_-_111330224
Show fit
|
0.06 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2
|
|
chr1_+_206635521
Show fit
|
0.06 |
ENST00000367109.8
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3
|
|
chr1_-_161238163
Show fit
|
0.06 |
ENST00000367982.8
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3
|
|
chr3_+_141262614
Show fit
|
0.06 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1
|
|
chr18_-_55402187
Show fit
|
0.06 |
ENST00000630268.2
ENST00000570177.6
|
TCF4
|
transcription factor 4
|
|
chr3_+_178419123
Show fit
|
0.06 |
ENST00000614557.1
ENST00000455307.5
ENST00000436432.1
|
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript)
long intergenic non-protein coding RNA 1014
|
|
chr13_+_24270681
Show fit
|
0.06 |
ENST00000343003.10
ENST00000399949.6
|
SPATA13
|
spermatogenesis associated 13
|
|
chr19_-_3500664
Show fit
|
0.06 |
ENST00000427575.6
|
DOHH
|
deoxyhypusine hydroxylase
|
|
chr3_+_101827982
Show fit
|
0.06 |
ENST00000461724.5
ENST00000483180.5
ENST00000394054.6
|
NFKBIZ
|
NFKB inhibitor zeta
|
|
chr4_+_87975667
Show fit
|
0.05 |
ENST00000237623.11
ENST00000682655.1
ENST00000508233.6
ENST00000360804.4
ENST00000395080.8
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_-_28058087
Show fit
|
0.05 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3
|
|
chr6_-_6004040
Show fit
|
0.05 |
ENST00000622188.4
|
NRN1
|
neuritin 1
|
|
chr4_-_122456725
Show fit
|
0.05 |
ENST00000226730.5
|
IL2
|
interleukin 2
|
|
chr2_-_157325808
Show fit
|
0.05 |
ENST00000410096.6
ENST00000420719.6
ENST00000409216.5
ENST00000419116.2
|
ERMN
|
ermin
|
|
chr4_+_87608529
Show fit
|
0.05 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein
|
|
chrX_+_41689006
Show fit
|
0.05 |
ENST00000378138.5
ENST00000620846.1
ENST00000649219.1
|
GPR34
|
G protein-coupled receptor 34
|
|
chr1_+_248038172
Show fit
|
0.05 |
ENST00000366479.4
|
OR2L2
|
olfactory receptor family 2 subfamily L member 2
|
|
chr16_-_48247533
Show fit
|
0.05 |
ENST00000356608.7
ENST00000569991.1
|
ABCC11
|
ATP binding cassette subfamily C member 11
|
|
chr16_+_76277393
Show fit
|
0.05 |
ENST00000611870.5
|
CNTNAP4
|
contactin associated protein family member 4
|
|
chrX_-_15600953
Show fit
|
0.05 |
ENST00000679212.1
ENST00000679278.1
ENST00000678046.1
ENST00000252519.8
|
ACE2
|
angiotensin I converting enzyme 2
|
|
chr19_-_3500625
Show fit
|
0.05 |
ENST00000672935.1
|
DOHH
|
deoxyhypusine hydroxylase
|
|
chr11_+_55635113
Show fit
|
0.05 |
ENST00000641760.1
|
OR4P4
|
olfactory receptor family 4 subfamily P member 4
|
|
chr7_+_16661182
Show fit
|
0.05 |
ENST00000446596.5
ENST00000452975.6
ENST00000438834.5
|
BZW2
|
basic leucine zipper and W2 domains 2
|
|
chr13_-_103066411
Show fit
|
0.05 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2
|
|
chr2_-_70835808
Show fit
|
0.05 |
ENST00000410009.5
|
CD207
|
CD207 molecule
|
|
chr4_+_94974984
Show fit
|
0.05 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr1_+_220094086
Show fit
|
0.05 |
ENST00000366922.3
|
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial
|
|
chr20_-_51768327
Show fit
|
0.05 |
ENST00000311637.9
ENST00000338821.6
|
ATP9A
|
ATPase phospholipid transporting 9A (putative)
|
|
chr2_+_9961165
Show fit
|
0.05 |
ENST00000405379.6
|
GRHL1
|
grainyhead like transcription factor 1
|
|
chr7_+_130344810
Show fit
|
0.05 |
ENST00000497503.5
ENST00000463587.5
ENST00000461828.5
ENST00000474905.6
ENST00000494311.1
ENST00000466363.6
|
CPA5
|
carboxypeptidase A5
|
|
chr6_+_44247866
Show fit
|
0.05 |
ENST00000371554.2
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1
|
|
chr9_-_101442403
Show fit
|
0.05 |
ENST00000648758.1
|
ALDOB
|
aldolase, fructose-bisphosphate B
|
|
chr1_+_44405164
Show fit
|
0.05 |
ENST00000355387.6
|
RNF220
|
ring finger protein 220
|
|
chr3_-_139677718
Show fit
|
0.05 |
ENST00000514703.5
ENST00000511444.5
ENST00000642987.1
ENST00000296202.11
ENST00000509291.5
ENST00000413939.6
ENST00000643695.2
ENST00000339837.9
ENST00000512391.5
ENST00000645507.1
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr15_-_55249029
Show fit
|
0.05 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr8_-_26867267
Show fit
|
0.05 |
ENST00000380573.4
|
ADRA1A
|
adrenoceptor alpha 1A
|
|
chr3_-_139678011
Show fit
|
0.05 |
ENST00000646611.1
ENST00000645290.1
ENST00000647257.1
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr9_-_101383558
Show fit
|
0.05 |
ENST00000674556.1
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase
|
|
chr1_+_92168915
Show fit
|
0.05 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8
|
|
chrY_+_18546691
Show fit
|
0.05 |
ENST00000309834.8
ENST00000307393.3
ENST00000382856.2
|
HSFY1
|
heat shock transcription factor Y-linked 1
|
|
chr3_-_129893551
Show fit
|
0.04 |
ENST00000505616.5
ENST00000426664.6
ENST00000648771.1
ENST00000393238.8
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr12_+_75334655
Show fit
|
0.04 |
ENST00000378695.9
|
GLIPR1L1
|
GLIPR1 like 1
|
|
chrY_-_18773686
Show fit
|
0.04 |
ENST00000382852.1
ENST00000344884.4
ENST00000304790.3
|
HSFY2
|
heat shock transcription factor Y-linked 2
|
|
chr9_+_74497308
Show fit
|
0.04 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B
|
|
chr13_-_46438190
Show fit
|
0.04 |
ENST00000409879.6
|
RUBCNL
|
rubicon like autophagy enhancer
|
|
chr2_-_208146150
Show fit
|
0.04 |
ENST00000260988.5
|
CRYGB
|
crystallin gamma B
|
|
chr6_-_29368865
Show fit
|
0.04 |
ENST00000641768.1
|
OR5V1
|
olfactory receptor family 5 subfamily V member 1
|
|
chr12_+_12357418
Show fit
|
0.04 |
ENST00000298571.6
|
BORCS5
|
BLOC-1 related complex subunit 5
|
|
chr12_+_75334675
Show fit
|
0.04 |
ENST00000312442.2
|
GLIPR1L1
|
GLIPR1 like 1
|
|
chr2_-_164773733
Show fit
|
0.04 |
ENST00000480873.1
|
COBLL1
|
cordon-bleu WH2 repeat protein like 1
|
|
chrX_-_135781729
Show fit
|
0.04 |
ENST00000617203.1
|
CT45A5
|
cancer/testis antigen family 45 member A5
|
|
chr1_-_152414256
Show fit
|
0.04 |
ENST00000271835.3
|
CRNN
|
cornulin
|
|
chr9_+_122371036
Show fit
|
0.04 |
ENST00000619306.5
ENST00000426608.6
ENST00000223423.8
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chrX_-_54043927
Show fit
|
0.04 |
ENST00000415025.5
ENST00000338946.10
|
PHF8
|
PHD finger protein 8
|
|
chr9_+_101185029
Show fit
|
0.04 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1
|
|
chr15_+_40594241
Show fit
|
0.04 |
ENST00000532056.5
ENST00000527044.5
ENST00000399668.7
|
KNL1
|
kinetochore scaffold 1
|
|
chr7_-_25228485
Show fit
|
0.04 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor
|
|
chr4_+_36281591
Show fit
|
0.04 |
ENST00000639862.2
ENST00000357504.7
|
DTHD1
|
death domain containing 1
|
|
chr8_-_3409528
Show fit
|
0.04 |
ENST00000335551.11
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr1_-_161238196
Show fit
|
0.04 |
ENST00000367983.9
ENST00000506209.5
ENST00000367980.6
ENST00000628566.2
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3
|
|
chr10_+_100347225
Show fit
|
0.04 |
ENST00000370355.3
|
SCD
|
stearoyl-CoA desaturase
|
|
chr7_+_107583919
Show fit
|
0.04 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29
|
|
chr12_+_54284258
Show fit
|
0.04 |
ENST00000677666.1
|
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1
|
|
chr6_+_158536398
Show fit
|
0.04 |
ENST00000367090.4
|
TMEM181
|
transmembrane protein 181
|
|
chrX_+_37780049
Show fit
|
0.04 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain
|
|
chr17_-_76570544
Show fit
|
0.03 |
ENST00000640006.1
|
ENSG00000284526.1
|
novel protein
|
|
chr5_+_148202771
Show fit
|
0.03 |
ENST00000514389.5
ENST00000621437.4
|
SPINK6
|
serine peptidase inhibitor Kazal type 6
|
|
chr1_-_44788168
Show fit
|
0.03 |
ENST00000372207.4
|
BEST4
|
bestrophin 4
|
|
chr16_-_21652598
Show fit
|
0.03 |
ENST00000569602.1
ENST00000268389.6
|
IGSF6
|
immunoglobulin superfamily member 6
|
|
chr17_-_10657302
Show fit
|
0.03 |
ENST00000583535.6
|
MYH3
|
myosin heavy chain 3
|
|
chr6_+_118548289
Show fit
|
0.03 |
ENST00000357525.6
|
PLN
|
phospholamban
|
|
chr10_-_88851809
Show fit
|
0.03 |
ENST00000371930.5
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr21_+_33025927
Show fit
|
0.03 |
ENST00000430860.1
ENST00000382357.4
ENST00000333337.3
|
OLIG2
|
oligodendrocyte transcription factor 2
|
|
chr19_+_36100593
Show fit
|
0.03 |
ENST00000680211.1
|
WDR62
|
WD repeat domain 62
|
|
chr14_+_30577752
Show fit
|
0.03 |
ENST00000547532.5
ENST00000555429.1
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr9_+_127264740
Show fit
|
0.03 |
ENST00000373387.9
|
GARNL3
|
GTPase activating Rap/RanGAP domain like 3
|
|
chr3_+_125969172
Show fit
|
0.03 |
ENST00000514116.6
ENST00000513830.5
|
ROPN1B
|
rhophilin associated tail protein 1B
|
|
chr2_+_32165841
Show fit
|
0.03 |
ENST00000357055.7
ENST00000435660.5
ENST00000440718.5
ENST00000379343.6
ENST00000282587.9
ENST00000406369.2
|
SLC30A6
|
solute carrier family 30 member 6
|
|
chr9_-_111759508
Show fit
|
0.03 |
ENST00000394777.8
ENST00000394779.7
|
SHOC1
|
shortage in chiasmata 1
|
|
chr13_-_79405784
Show fit
|
0.03 |
ENST00000267229.11
|
RBM26
|
RNA binding motif protein 26
|
|
chr14_+_56117702
Show fit
|
0.03 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2
|
|
chrX_-_45200895
Show fit
|
0.03 |
ENST00000377934.4
|
DIPK2B
|
divergent protein kinase domain 2B
|
|
chr16_+_56191476
Show fit
|
0.03 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1
|
|
chr6_+_26156323
Show fit
|
0.03 |
ENST00000304218.6
|
H1-4
|
H1.4 linker histone, cluster member
|
|
chrX_-_123623155
Show fit
|
0.03 |
ENST00000618150.4
|
THOC2
|
THO complex 2
|
|
chr11_+_60280658
Show fit
|
0.03 |
ENST00000337908.5
|
MS4A4A
|
membrane spanning 4-domains A4A
|
|
chr9_-_101435760
Show fit
|
0.03 |
ENST00000647789.2
ENST00000616752.1
|
ALDOB
|
aldolase, fructose-bisphosphate B
|
|
chr11_-_119101814
Show fit
|
0.03 |
ENST00000682791.1
ENST00000639704.1
ENST00000354202.9
|
DPAGT1
|
dolichyl-phosphate N-acetylglucosaminephosphotransferase 1
|
|
chr4_+_154563003
Show fit
|
0.03 |
ENST00000302068.9
ENST00000509493.1
|
FGB
|
fibrinogen beta chain
|
|
chr15_-_58279245
Show fit
|
0.03 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2
|
|
chr9_+_60914407
Show fit
|
0.03 |
ENST00000437823.5
|
SPATA31A5
|
SPATA31 subfamily A member 5
|
|
chr4_+_71062642
Show fit
|
0.03 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4
|
|
chr3_+_172754457
Show fit
|
0.03 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2
|
|
chr20_-_7940444
Show fit
|
0.03 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1
|
|
chr12_+_16347102
Show fit
|
0.03 |
ENST00000536371.5
ENST00000010404.6
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr12_-_21775581
Show fit
|
0.03 |
ENST00000537950.1
ENST00000665145.1
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8
|
|
chr12_-_118190510
Show fit
|
0.03 |
ENST00000540561.5
ENST00000537952.1
ENST00000537822.1
|
TAOK3
|
TAO kinase 3
|
|
chr8_+_131939865
Show fit
|
0.03 |
ENST00000520362.5
ENST00000519656.1
|
EFR3A
|
EFR3 homolog A
|
|
chr2_+_28496054
Show fit
|
0.03 |
ENST00000327757.10
ENST00000422425.6
ENST00000404858.5
|
PLB1
|
phospholipase B1
|
|
chr1_-_161238085
Show fit
|
0.03 |
ENST00000512372.5
ENST00000437437.6
ENST00000412844.6
ENST00000428574.6
ENST00000442691.6
ENST00000505005.5
ENST00000508740.5
ENST00000367981.7
ENST00000502985.5
ENST00000504010.5
ENST00000508387.5
ENST00000511676.5
ENST00000511748.5
ENST00000511944.5
ENST00000515621.5
ENST00000367984.8
ENST00000367985.7
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3
|
|
chr19_+_48393657
Show fit
|
0.03 |
ENST00000263269.4
|
GRIN2D
|
glutamate ionotropic receptor NMDA type subunit 2D
|
|
chr2_-_162536955
Show fit
|
0.03 |
ENST00000621889.1
|
KCNH7
|
potassium voltage-gated channel subfamily H member 7
|
|
chr12_-_95551417
Show fit
|
0.03 |
ENST00000258499.8
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr13_+_45464901
Show fit
|
0.02 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3
|