Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
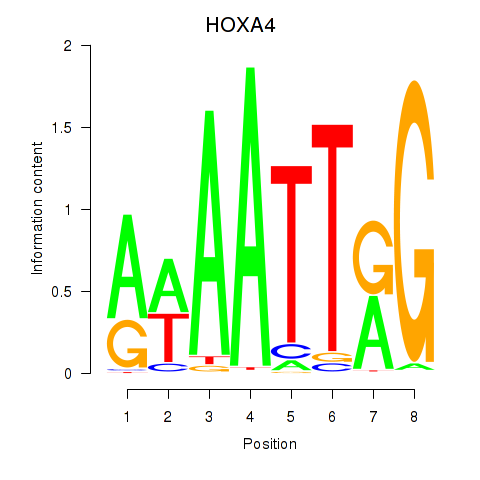
Results for HOXA4
Z-value: 0.71
Transcription factors associated with HOXA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA4
|
ENSG00000197576.14 | homeobox A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA4 | hg38_v1_chr7_-_27130182_27130226 | -0.55 | 1.6e-01 | Click! |
Activity profile of HOXA4 motif
Sorted Z-values of HOXA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_104085847 | 1.81 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr5_+_53480619 | 1.56 |
ENST00000396947.7
ENST00000256759.8 |
FST
|
follistatin |
| chr5_-_1882902 | 0.94 |
ENST00000231357.7
|
IRX4
|
iroquois homeobox 4 |
| chr5_+_67004618 | 0.78 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_-_53956811 | 0.60 |
ENST00000559087.5
ENST00000245451.9 |
BMP4
|
bone morphogenetic protein 4 |
| chr4_+_83536097 | 0.56 |
ENST00000395226.6
ENST00000264409.5 |
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr18_+_31376777 | 0.55 |
ENST00000308128.9
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr15_+_90201301 | 0.53 |
ENST00000411539.6
|
SEMA4B
|
semaphorin 4B |
| chr8_-_42501224 | 0.52 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr1_-_204359885 | 0.43 |
ENST00000414478.1
ENST00000272203.8 |
PLEKHA6
|
pleckstrin homology domain containing A6 |
| chrX_-_85379659 | 0.43 |
ENST00000262753.9
|
POF1B
|
POF1B actin binding protein |
| chr6_+_121437378 | 0.41 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr11_+_112175526 | 0.39 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr12_+_29223730 | 0.38 |
ENST00000547116.5
|
FAR2
|
fatty acyl-CoA reductase 2 |
| chr12_+_29223659 | 0.38 |
ENST00000182377.8
|
FAR2
|
fatty acyl-CoA reductase 2 |
| chr17_-_41382298 | 0.37 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
| chr1_+_81306096 | 0.37 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr14_-_37595224 | 0.36 |
ENST00000250448.5
|
FOXA1
|
forkhead box A1 |
| chr11_+_112175501 | 0.36 |
ENST00000357685.11
ENST00000361053.8 |
BCO2
|
beta-carotene oxygenase 2 |
| chr13_-_46142834 | 0.35 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr1_-_216805367 | 0.35 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr2_-_68871382 | 0.33 |
ENST00000295379.2
|
BMP10
|
bone morphogenetic protein 10 |
| chr18_-_55351977 | 0.32 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr18_-_55422492 | 0.31 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr5_+_148203024 | 0.30 |
ENST00000325630.3
|
SPINK6
|
serine peptidase inhibitor Kazal type 6 |
| chr11_+_128694052 | 0.30 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_+_128693887 | 0.29 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_38557459 | 0.29 |
ENST00000511561.1
|
LIFR
|
LIF receptor subunit alpha |
| chr5_+_148202771 | 0.29 |
ENST00000514389.5
ENST00000621437.4 |
SPINK6
|
serine peptidase inhibitor Kazal type 6 |
| chr7_+_144355288 | 0.28 |
ENST00000498580.5
ENST00000056217.10 |
ARHGEF5
|
Rho guanine nucleotide exchange factor 5 |
| chr12_-_23951020 | 0.28 |
ENST00000441133.2
ENST00000545921.5 |
SOX5
|
SRY-box transcription factor 5 |
| chr7_+_95485898 | 0.27 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chrX_-_120311533 | 0.27 |
ENST00000440464.5
ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr18_-_31162849 | 0.27 |
ENST00000257197.7
ENST00000257198.6 |
DSC1
|
desmocollin 1 |
| chr11_-_16356538 | 0.26 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chrX_-_120311452 | 0.26 |
ENST00000371369.9
|
TMEM255A
|
transmembrane protein 255A |
| chrX_-_120311408 | 0.25 |
ENST00000309720.9
|
TMEM255A
|
transmembrane protein 255A |
| chr20_+_54475584 | 0.24 |
ENST00000262593.10
|
DOK5
|
docking protein 5 |
| chrX_+_135520616 | 0.24 |
ENST00000370752.4
ENST00000639893.2 |
INTS6L
|
integrator complex subunit 6 like |
| chr11_-_59615673 | 0.23 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein |
| chr2_+_127645864 | 0.23 |
ENST00000544369.5
|
GPR17
|
G protein-coupled receptor 17 |
| chr17_+_39738317 | 0.22 |
ENST00000394211.7
|
GRB7
|
growth factor receptor bound protein 7 |
| chr8_-_94436926 | 0.21 |
ENST00000481490.3
|
FSBP
|
fibrinogen silencer binding protein |
| chr18_+_44680093 | 0.21 |
ENST00000426838.8
ENST00000677068.1 |
SETBP1
|
SET binding protein 1 |
| chr18_-_26548527 | 0.21 |
ENST00000580059.7
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr1_+_160190567 | 0.20 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr14_+_39265278 | 0.20 |
ENST00000554392.5
ENST00000555716.5 ENST00000341749.7 ENST00000557038.5 |
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr2_+_127646145 | 0.19 |
ENST00000486700.2
ENST00000272644.7 |
GPR17
|
G protein-coupled receptor 17 |
| chr17_-_50707855 | 0.19 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40 |
| chrX_-_85379703 | 0.19 |
ENST00000373145.3
|
POF1B
|
POF1B actin binding protein |
| chr19_-_1021114 | 0.18 |
ENST00000333175.9
ENST00000356663.8 |
TMEM259
|
transmembrane protein 259 |
| chr2_+_176151543 | 0.17 |
ENST00000306324.4
|
HOXD4
|
homeobox D4 |
| chr15_+_74788542 | 0.16 |
ENST00000567571.5
|
CSK
|
C-terminal Src kinase |
| chr4_-_152352800 | 0.16 |
ENST00000393956.9
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr14_+_45135917 | 0.15 |
ENST00000267430.10
ENST00000556036.5 ENST00000542564.6 |
FANCM
|
FA complementation group M |
| chrM_+_5824 | 0.15 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr2_+_170715317 | 0.15 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor |
| chr10_-_60572599 | 0.14 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chrX_-_123623155 | 0.14 |
ENST00000618150.4
|
THOC2
|
THO complex 2 |
| chr2_+_53767772 | 0.14 |
ENST00000295304.5
|
CHAC2
|
ChaC glutathione specific gamma-glutamylcyclotransferase 2 |
| chr19_+_49591170 | 0.14 |
ENST00000418929.7
|
PRR12
|
proline rich 12 |
| chr10_-_67696115 | 0.14 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr12_+_56128217 | 0.14 |
ENST00000267113.4
ENST00000394048.10 |
ESYT1
|
extended synaptotagmin 1 |
| chrX_+_37780049 | 0.14 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr3_-_52452828 | 0.13 |
ENST00000496590.1
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr12_+_54053815 | 0.13 |
ENST00000430889.3
|
HOXC4
|
homeobox C4 |
| chr16_+_7332839 | 0.13 |
ENST00000355637.9
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr5_+_66958870 | 0.12 |
ENST00000405643.5
ENST00000407621.1 ENST00000432426.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_95073490 | 0.12 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2 group C member 1 |
| chr17_-_40799939 | 0.12 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chrX_+_41689006 | 0.12 |
ENST00000378138.5
ENST00000620846.1 ENST00000649219.1 |
GPR34
|
G protein-coupled receptor 34 |
| chr3_-_120682215 | 0.12 |
ENST00000283871.10
|
HGD
|
homogentisate 1,2-dioxygenase |
| chrX_+_41688967 | 0.12 |
ENST00000378142.9
|
GPR34
|
G protein-coupled receptor 34 |
| chr17_+_42844573 | 0.12 |
ENST00000253799.8
ENST00000452774.2 |
AOC2
|
amine oxidase copper containing 2 |
| chr6_+_99606833 | 0.11 |
ENST00000369215.5
|
PRDM13
|
PR/SET domain 13 |
| chr7_+_95485934 | 0.11 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr6_+_12716554 | 0.11 |
ENST00000676159.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr9_+_84669760 | 0.11 |
ENST00000304053.10
|
NTRK2
|
neurotrophic receptor tyrosine kinase 2 |
| chr3_+_188947199 | 0.11 |
ENST00000433971.5
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr5_-_77638713 | 0.11 |
ENST00000306422.5
|
OTP
|
orthopedia homeobox |
| chr4_+_15427998 | 0.10 |
ENST00000444304.3
|
C1QTNF7
|
C1q and TNF related 7 |
| chr17_-_10518536 | 0.10 |
ENST00000226207.6
|
MYH1
|
myosin heavy chain 1 |
| chr17_-_10549652 | 0.10 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr5_-_88883701 | 0.10 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr6_+_12716801 | 0.10 |
ENST00000674595.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr6_+_12716760 | 0.10 |
ENST00000332995.12
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr6_+_32439866 | 0.10 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr3_+_121593363 | 0.09 |
ENST00000338040.6
|
FBXO40
|
F-box protein 40 |
| chr17_-_10549612 | 0.09 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
| chr1_+_155689074 | 0.09 |
ENST00000343043.7
ENST00000421487.6 ENST00000535183.5 ENST00000368336.10 ENST00000465375.5 ENST00000470830.5 |
DAP3
|
death associated protein 3 |
| chr18_-_23437927 | 0.09 |
ENST00000578520.5
ENST00000383233.8 |
TMEM241
|
transmembrane protein 241 |
| chr8_-_10655137 | 0.09 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chr12_-_86256267 | 0.09 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr14_-_36519679 | 0.09 |
ENST00000498187.6
|
NKX2-1
|
NK2 homeobox 1 |
| chr17_-_10549694 | 0.09 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chr14_+_22515623 | 0.09 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr16_+_7332744 | 0.08 |
ENST00000436368.6
ENST00000311745.9 ENST00000340209.8 ENST00000620507.4 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr3_+_122384167 | 0.08 |
ENST00000232125.9
ENST00000477892.5 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162 member A |
| chr17_-_48593748 | 0.08 |
ENST00000239151.6
|
HOXB5
|
homeobox B5 |
| chr7_+_107470050 | 0.08 |
ENST00000304402.6
|
GPR22
|
G protein-coupled receptor 22 |
| chr11_-_69819410 | 0.08 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr2_-_179562576 | 0.08 |
ENST00000336917.9
|
ZNF385B
|
zinc finger protein 385B |
| chr12_+_49750598 | 0.08 |
ENST00000552370.5
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr6_+_50818857 | 0.08 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr6_-_55875583 | 0.08 |
ENST00000370830.4
|
BMP5
|
bone morphogenetic protein 5 |
| chr11_+_27041313 | 0.08 |
ENST00000528583.5
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr5_+_55853464 | 0.08 |
ENST00000652039.1
|
IL31RA
|
interleukin 31 receptor A |
| chrX_-_111410420 | 0.07 |
ENST00000371993.7
ENST00000680476.1 |
DCX
|
doublecortin |
| chr10_+_113125536 | 0.07 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr2_-_80304700 | 0.07 |
ENST00000295057.4
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr1_-_156490599 | 0.07 |
ENST00000360595.7
|
MEF2D
|
myocyte enhancer factor 2D |
| chr20_+_54475647 | 0.07 |
ENST00000395939.5
|
DOK5
|
docking protein 5 |
| chr9_+_84669707 | 0.07 |
ENST00000277120.8
ENST00000323115.10 |
NTRK2
|
neurotrophic receptor tyrosine kinase 2 |
| chr12_+_53425070 | 0.07 |
ENST00000550839.1
|
AMHR2
|
anti-Mullerian hormone receptor type 2 |
| chr13_-_83882390 | 0.07 |
ENST00000377084.3
|
SLITRK1
|
SLIT and NTRK like family member 1 |
| chr6_+_151690492 | 0.07 |
ENST00000404742.5
ENST00000440973.5 |
ESR1
|
estrogen receptor 1 |
| chr2_-_80304274 | 0.06 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr1_+_167329044 | 0.06 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1 |
| chr13_-_83882456 | 0.06 |
ENST00000674365.1
|
SLITRK1
|
SLIT and NTRK like family member 1 |
| chr12_+_71664352 | 0.06 |
ENST00000547843.1
|
THAP2
|
THAP domain containing 2 |
| chr1_-_100178215 | 0.06 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr6_-_116126120 | 0.06 |
ENST00000452729.1
ENST00000651968.1 ENST00000243222.8 |
COL10A1
|
collagen type X alpha 1 chain |
| chr2_-_2324323 | 0.06 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr13_-_52848632 | 0.05 |
ENST00000377942.7
ENST00000338862.5 |
PCDH8
|
protocadherin 8 |
| chrX_-_54998530 | 0.05 |
ENST00000545676.5
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr12_-_95073580 | 0.05 |
ENST00000393101.7
ENST00000333003.10 ENST00000622476.4 |
NR2C1
|
nuclear receptor subfamily 2 group C member 1 |
| chr7_+_114416286 | 0.05 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2 |
| chr11_+_22666604 | 0.04 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr3_+_52414523 | 0.04 |
ENST00000461861.5
|
PHF7
|
PHD finger protein 7 |
| chr14_-_20436161 | 0.04 |
ENST00000636854.3
|
KLHL33
|
kelch like family member 33 |
| chr8_-_28490220 | 0.04 |
ENST00000517673.5
ENST00000380254.7 ENST00000518734.5 ENST00000346498.6 |
FBXO16
|
F-box protein 16 |
| chr8_-_66178455 | 0.03 |
ENST00000276571.5
|
CRH
|
corticotropin releasing hormone |
| chr19_+_33373694 | 0.03 |
ENST00000284000.9
|
CEBPG
|
CCAAT enhancer binding protein gamma |
| chr7_+_129375643 | 0.03 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr9_+_92964272 | 0.03 |
ENST00000468206.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr3_+_107523026 | 0.03 |
ENST00000416476.6
|
BBX
|
BBX high mobility group box domain containing |
| chr5_+_153490727 | 0.03 |
ENST00000340592.9
|
GRIA1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chr12_+_106582996 | 0.02 |
ENST00000392842.6
|
RFX4
|
regulatory factor X4 |
| chr5_+_70025247 | 0.02 |
ENST00000380751.9
ENST00000380750.8 ENST00000503931.5 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B |
| chr8_-_98942557 | 0.02 |
ENST00000523601.5
|
STK3
|
serine/threonine kinase 3 |
| chr10_-_73655984 | 0.02 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like |
| chr4_-_184217854 | 0.02 |
ENST00000296741.7
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr18_+_3247778 | 0.02 |
ENST00000217652.8
ENST00000578611.5 ENST00000583449.1 |
MYL12A
|
myosin light chain 12A |
| chr11_+_92224801 | 0.02 |
ENST00000525166.6
|
FAT3
|
FAT atypical cadherin 3 |
| chr2_-_190062721 | 0.02 |
ENST00000260950.5
|
MSTN
|
myostatin |
| chr10_+_116545907 | 0.02 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr10_-_119178791 | 0.02 |
ENST00000298510.4
|
PRDX3
|
peroxiredoxin 3 |
| chr15_+_36702009 | 0.02 |
ENST00000562489.1
|
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr5_+_141430565 | 0.01 |
ENST00000613314.1
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr7_+_117611617 | 0.01 |
ENST00000468795.1
|
CFTR
|
CF transmembrane conductance regulator |
| chr7_-_141014629 | 0.01 |
ENST00000393008.7
|
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr12_-_120369156 | 0.01 |
ENST00000257552.7
|
MSI1
|
musashi RNA binding protein 1 |
| chr1_+_213987929 | 0.01 |
ENST00000498508.6
ENST00000366958.9 |
PROX1
|
prospero homeobox 1 |
| chr2_+_167187364 | 0.01 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr17_-_10421944 | 0.01 |
ENST00000403437.2
|
MYH8
|
myosin heavy chain 8 |
| chr20_+_33562306 | 0.01 |
ENST00000344201.7
|
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr1_+_213988501 | 0.01 |
ENST00000261454.8
ENST00000435016.2 |
PROX1
|
prospero homeobox 1 |
| chr1_+_26021768 | 0.01 |
ENST00000374280.4
|
EXTL1
|
exostosin like glycosyltransferase 1 |
| chr7_+_32957385 | 0.01 |
ENST00000538336.5
ENST00000242209.9 |
FKBP9
|
FKBP prolyl isomerase 9 |
| chr12_-_56643499 | 0.01 |
ENST00000551570.5
|
ATP5F1B
|
ATP synthase F1 subunit beta |
| chr17_+_56153458 | 0.00 |
ENST00000318698.6
ENST00000682825.1 ENST00000566473.6 |
ANKFN1
|
ankyrin repeat and fibronectin type III domain containing 1 |
| chr4_+_87832917 | 0.00 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr3_+_107522936 | 0.00 |
ENST00000415149.6
ENST00000402543.5 ENST00000325805.13 ENST00000427402.5 |
BBX
|
BBX high mobility group box domain containing |
| chr1_+_28259473 | 0.00 |
ENST00000253063.4
|
SESN2
|
sestrin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.2 | 0.7 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.2 | 0.6 | GO:0072099 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.8 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.4 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.4 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 1.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 0.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.2 | GO:0014894 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.0 | 1.9 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) negative regulation of glucagon secretion(GO:0070093) cellular response to cocaine(GO:0071314) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 1.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |