Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
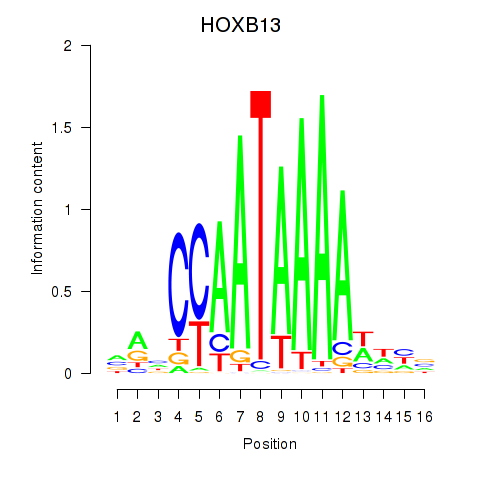
Results for HOXB13
Z-value: 0.45
Transcription factors associated with HOXB13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB13
|
ENSG00000159184.8 | homeobox B13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB13 | hg38_v1_chr17_-_48728705_48728756 | 0.26 | 5.3e-01 | Click! |
Activity profile of HOXB13 motif
Sorted Z-values of HOXB13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_78303865 | 0.84 |
ENST00000370758.5
|
PTGFR
|
prostaglandin F receptor |
| chr2_-_189179754 | 0.82 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr8_+_96584920 | 0.79 |
ENST00000521590.5
|
SDC2
|
syndecan 2 |
| chr3_-_195583931 | 0.33 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr11_-_102955705 | 0.32 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr8_+_76681208 | 0.28 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr3_-_115071333 | 0.27 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr14_-_50561119 | 0.24 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_+_244352627 | 0.21 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr17_+_18183052 | 0.18 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr3_-_15797930 | 0.18 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr5_+_136160986 | 0.17 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr7_-_75772172 | 0.17 |
ENST00000005180.9
|
CCL26
|
C-C motif chemokine ligand 26 |
| chr9_+_122371014 | 0.15 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr16_+_30378312 | 0.13 |
ENST00000528032.5
ENST00000622647.3 |
ZNF48
|
zinc finger protein 48 |
| chr2_+_74458400 | 0.13 |
ENST00000393972.7
ENST00000233615.7 ENST00000409737.5 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr21_-_34526850 | 0.12 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr9_-_128067310 | 0.12 |
ENST00000373078.5
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr20_+_58907981 | 0.12 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr3_-_15798184 | 0.11 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr12_+_8157034 | 0.10 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr16_-_24930952 | 0.09 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr1_-_155910881 | 0.09 |
ENST00000609492.1
ENST00000368322.7 |
RIT1
|
Ras like without CAAX 1 |
| chr16_-_24931090 | 0.09 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr6_+_122471913 | 0.09 |
ENST00000615438.4
ENST00000392491.6 |
PKIB
|
cAMP-dependent protein kinase inhibitor beta |
| chr19_-_5903703 | 0.08 |
ENST00000586349.5
ENST00000585661.1 ENST00000592634.5 ENST00000418389.6 ENST00000308961.5 |
ENSG00000267740.5
NDUFA11
|
novel protein NADH:ubiquinone oxidoreductase subunit A11 |
| chr2_-_174764407 | 0.08 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr1_+_84164962 | 0.08 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_-_123991352 | 0.08 |
ENST00000184183.8
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr1_+_210328244 | 0.07 |
ENST00000541565.5
ENST00000413764.6 |
HHAT
|
hedgehog acyltransferase |
| chr2_-_135530561 | 0.07 |
ENST00000536680.5
ENST00000401392.5 |
ZRANB3
|
zinc finger RANBP2-type containing 3 |
| chr6_-_43053832 | 0.07 |
ENST00000265348.9
ENST00000674134.1 ENST00000674100.1 |
CUL7
|
cullin 7 |
| chr14_-_77423517 | 0.06 |
ENST00000555603.1
|
NOXRED1
|
NADP dependent oxidoreductase domain containing 1 |
| chr1_-_36397880 | 0.06 |
ENST00000315732.3
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr17_+_18183803 | 0.05 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr1_-_45491150 | 0.05 |
ENST00000372086.4
|
TESK2
|
testis associated actin remodelling kinase 2 |
| chr5_-_145835330 | 0.05 |
ENST00000394450.6
ENST00000683046.1 |
PRELID2
|
PRELI domain containing 2 |
| chrM_-_14669 | 0.05 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 |
| chr7_+_133253064 | 0.05 |
ENST00000393161.6
ENST00000253861.5 |
EXOC4
|
exocyst complex component 4 |
| chr22_+_35066136 | 0.04 |
ENST00000308700.6
ENST00000404699.7 |
ISX
|
intestine specific homeobox |
| chr11_-_102843597 | 0.04 |
ENST00000299855.10
|
MMP3
|
matrix metallopeptidase 3 |
| chr9_-_14180779 | 0.04 |
ENST00000380924.1
ENST00000543693.5 |
NFIB
|
nuclear factor I B |
| chr1_+_84164370 | 0.04 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr5_+_102808057 | 0.04 |
ENST00000684043.1
ENST00000682407.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr6_-_27814757 | 0.04 |
ENST00000333151.5
|
H2AC14
|
H2A clustered histone 14 |
| chr19_+_47994696 | 0.03 |
ENST00000596043.5
ENST00000597519.5 |
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr9_+_122370523 | 0.03 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr17_-_48728705 | 0.03 |
ENST00000290295.8
|
HOXB13
|
homeobox B13 |
| chr19_+_47994625 | 0.03 |
ENST00000339841.7
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr18_+_3252267 | 0.03 |
ENST00000536605.1
ENST00000580887.5 |
MYL12A
|
myosin light chain 12A |
| chr16_-_71577082 | 0.03 |
ENST00000355962.5
|
TAT
|
tyrosine aminotransferase |
| chr2_-_174764436 | 0.03 |
ENST00000409323.1
ENST00000261007.9 ENST00000348749.9 ENST00000672640.1 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr4_+_67558719 | 0.03 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr9_+_101185029 | 0.03 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr20_-_10420737 | 0.03 |
ENST00000649912.1
|
ENSG00000285723.1
|
novel protein |
| chr5_-_145835285 | 0.02 |
ENST00000505416.5
ENST00000334744.8 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr1_+_44405164 | 0.02 |
ENST00000355387.6
|
RNF220
|
ring finger protein 220 |
| chr5_-_127073467 | 0.02 |
ENST00000607731.5
ENST00000509733.7 ENST00000296662.10 ENST00000535381.6 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr5_-_140673568 | 0.02 |
ENST00000542735.2
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr14_-_67533720 | 0.02 |
ENST00000554278.6
|
TMEM229B
|
transmembrane protein 229B |
| chr19_+_55643592 | 0.02 |
ENST00000270451.6
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr1_-_149812359 | 0.02 |
ENST00000369167.2
ENST00000545683.1 |
H2BC18
|
H2B clustered histone 18 |
| chr9_+_122371036 | 0.02 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_73151833 | 0.01 |
ENST00000456643.5
ENST00000257497.11 ENST00000415424.5 |
ANXA1
|
annexin A1 |
| chr7_-_11832190 | 0.01 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr6_+_27815010 | 0.01 |
ENST00000621112.2
|
H2BC14
|
H2B clustered histone 14 |
| chr15_-_55249029 | 0.01 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_+_109827963 | 0.01 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr12_-_81598332 | 0.01 |
ENST00000443686.7
|
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr2_-_175181663 | 0.01 |
ENST00000392541.3
ENST00000284727.9 ENST00000409194.5 |
ATP5MC3
|
ATP synthase membrane subunit c locus 3 |
| chr1_+_50105666 | 0.00 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr12_-_52321395 | 0.00 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr18_+_3252208 | 0.00 |
ENST00000578562.6
|
MYL12A
|
myosin light chain 12A |
| chr22_-_37844308 | 0.00 |
ENST00000411961.6
ENST00000434930.1 ENST00000215941.9 |
ANKRD54
|
ankyrin repeat domain 54 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.8 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.2 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |