Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
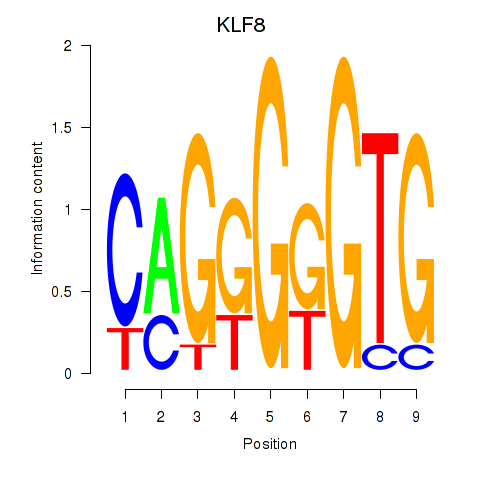
Results for KLF8
Z-value: 0.80
Transcription factors associated with KLF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF8
|
ENSG00000102349.18 | Kruppel like factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF8 | hg38_v1_chrX_+_56232343_56232363 | 0.12 | 7.7e-01 | Click! |
Activity profile of KLF8 motif
Sorted Z-values of KLF8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_76403266 | 0.41 |
ENST00000274364.11
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr4_-_99352754 | 0.40 |
ENST00000639454.1
|
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr1_-_153544997 | 0.37 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr17_-_19745602 | 0.35 |
ENST00000444455.5
ENST00000439102.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr4_-_99321362 | 0.34 |
ENST00000625860.2
ENST00000305046.13 ENST00000506651.5 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr5_+_321695 | 0.32 |
ENST00000684583.1
ENST00000514523.1 |
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr11_-_1753870 | 0.31 |
ENST00000636579.1
|
ENSG00000250644.3
|
novel protein |
| chrX_+_16946862 | 0.29 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr14_-_91947654 | 0.28 |
ENST00000342058.9
|
FBLN5
|
fibulin 5 |
| chr6_+_84033717 | 0.28 |
ENST00000257776.5
|
MRAP2
|
melanocortin 2 receptor accessory protein 2 |
| chr11_+_86800507 | 0.28 |
ENST00000533902.2
|
PRSS23
|
serine protease 23 |
| chr1_-_3611470 | 0.27 |
ENST00000356575.9
|
MEGF6
|
multiple EGF like domains 6 |
| chr19_+_43581037 | 0.25 |
ENST00000562365.2
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr11_+_314010 | 0.25 |
ENST00000680699.1
ENST00000681938.1 ENST00000679380.1 ENST00000680938.1 ENST00000408968.4 ENST00000681180.1 ENST00000681426.1 ENST00000680696.1 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr3_-_58587033 | 0.25 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107 member A |
| chr8_+_103371490 | 0.24 |
ENST00000330295.10
ENST00000415886.2 |
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr20_+_64066211 | 0.24 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A2 |
| chr2_+_24123454 | 0.24 |
ENST00000615575.5
|
FAM228B
|
family with sequence similarity 228 member B |
| chr17_+_1762052 | 0.23 |
ENST00000254722.9
ENST00000576406.5 ENST00000571149.5 |
SERPINF1
|
serpin family F member 1 |
| chr14_+_105474781 | 0.22 |
ENST00000550577.5
ENST00000538259.2 ENST00000329146.9 |
CRIP2
|
cysteine rich protein 2 |
| chr17_-_19748355 | 0.22 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr1_-_153545793 | 0.21 |
ENST00000354332.8
ENST00000368716.9 |
S100A4
|
S100 calcium binding protein A4 |
| chr11_-_101129042 | 0.21 |
ENST00000263463.9
|
PGR
|
progesterone receptor |
| chr20_+_64063105 | 0.21 |
ENST00000395053.7
ENST00000343484.10 ENST00000339217.8 |
TCEA2
|
transcription elongation factor A2 |
| chr11_-_111912871 | 0.21 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr14_-_91946989 | 0.20 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr8_+_103372388 | 0.20 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr19_+_917287 | 0.20 |
ENST00000592648.1
ENST00000234371.10 |
KISS1R
|
KISS1 receptor |
| chr2_+_209424039 | 0.19 |
ENST00000682079.1
ENST00000199940.10 |
MAP2
|
microtubule associated protein 2 |
| chr6_-_31729260 | 0.19 |
ENST00000375789.7
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr21_-_6467509 | 0.19 |
ENST00000624406.3
ENST00000398168.5 ENST00000624934.3 |
CBSL
|
cystathionine beta-synthase like |
| chr14_+_73237493 | 0.19 |
ENST00000340738.9
ENST00000644200.2 |
PAPLN
|
papilin, proteoglycan like sulfated glycoprotein |
| chr11_+_117200188 | 0.19 |
ENST00000529792.5
|
TAGLN
|
transgelin |
| chr10_-_13300051 | 0.19 |
ENST00000479604.1
ENST00000263038.9 |
PHYH
|
phytanoyl-CoA 2-hydroxylase |
| chr16_+_67279508 | 0.19 |
ENST00000379344.8
ENST00000568621.1 ENST00000450733.5 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chr14_-_105940235 | 0.18 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr2_-_10080411 | 0.18 |
ENST00000381813.4
|
CYS1
|
cystin 1 |
| chr7_+_48455063 | 0.18 |
ENST00000411975.5
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr17_-_19745369 | 0.18 |
ENST00000573368.5
ENST00000457500.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr17_+_19648915 | 0.18 |
ENST00000672567.1
ENST00000672709.1 |
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr15_-_93073706 | 0.18 |
ENST00000425933.6
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr1_+_109712272 | 0.18 |
ENST00000369812.6
|
GSTM5
|
glutathione S-transferase mu 5 |
| chr2_+_24049705 | 0.17 |
ENST00000380986.9
ENST00000452109.1 |
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr19_+_12751789 | 0.17 |
ENST00000553030.6
|
BEST2
|
bestrophin 2 |
| chrX_+_16946650 | 0.17 |
ENST00000357277.8
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr2_+_127423265 | 0.17 |
ENST00000402125.2
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr9_-_92482350 | 0.17 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr20_+_13008919 | 0.17 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr1_+_78490966 | 0.17 |
ENST00000370757.8
ENST00000370756.3 |
PTGFR
|
prostaglandin F receptor |
| chr17_+_78232006 | 0.17 |
ENST00000550981.7
ENST00000591033.2 |
TMEM235
|
transmembrane protein 235 |
| chr19_+_15949008 | 0.17 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr22_+_18906303 | 0.17 |
ENST00000331444.12
ENST00000608842.1 |
DGCR6
|
DiGeorge syndrome critical region gene 6 |
| chr17_-_81937221 | 0.16 |
ENST00000402252.6
ENST00000583564.5 ENST00000585244.1 ENST00000337943.9 ENST00000579698.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr6_+_16238554 | 0.16 |
ENST00000259727.5
|
GMPR
|
guanosine monophosphate reductase |
| chr19_-_17303369 | 0.16 |
ENST00000247706.4
ENST00000594194.1 |
ABHD8
|
abhydrolase domain containing 8 |
| chr19_+_17226662 | 0.16 |
ENST00000598068.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr9_+_36036899 | 0.16 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr3_-_58627596 | 0.16 |
ENST00000474531.5
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107 member A |
| chr19_+_35031263 | 0.16 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr2_+_24049673 | 0.16 |
ENST00000380991.8
|
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr22_+_37019635 | 0.16 |
ENST00000401419.7
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr21_+_46098102 | 0.16 |
ENST00000300527.9
ENST00000310645.9 |
COL6A2
|
collagen type VI alpha 2 chain |
| chr8_+_22567038 | 0.15 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr9_-_92404559 | 0.15 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr11_+_308106 | 0.15 |
ENST00000399817.9
ENST00000680344.1 |
IFITM2
|
interferon induced transmembrane protein 2 |
| chr22_+_37019688 | 0.15 |
ENST00000404802.7
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr4_-_56656304 | 0.15 |
ENST00000503639.7
|
HOPX
|
HOP homeobox |
| chr9_-_97697297 | 0.15 |
ENST00000375128.5
|
XPA
|
XPA, DNA damage recognition and repair factor |
| chr15_+_32718476 | 0.15 |
ENST00000652365.1
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr10_-_91633057 | 0.15 |
ENST00000238994.6
|
PPP1R3C
|
protein phosphatase 1 regulatory subunit 3C |
| chr6_-_166627244 | 0.15 |
ENST00000265678.9
|
RPS6KA2
|
ribosomal protein S6 kinase A2 |
| chr19_+_43580544 | 0.15 |
ENST00000562255.5
ENST00000569031.6 |
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr12_-_108339300 | 0.15 |
ENST00000550402.6
ENST00000552995.5 ENST00000312143.11 |
CMKLR1
|
chemerin chemokine-like receptor 1 |
| chr12_+_51239278 | 0.15 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr11_+_46381194 | 0.15 |
ENST00000533952.5
|
MDK
|
midkine |
| chr1_+_215082731 | 0.15 |
ENST00000444842.7
|
KCNK2
|
potassium two pore domain channel subfamily K member 2 |
| chr16_+_2029636 | 0.15 |
ENST00000561844.5
|
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr22_+_37019735 | 0.15 |
ENST00000429360.6
ENST00000341116.7 ENST00000404393.5 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr17_-_81937277 | 0.14 |
ENST00000405481.8
ENST00000329875.13 ENST00000585215.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr19_-_54473208 | 0.14 |
ENST00000301200.3
|
CDC42EP5
|
CDC42 effector protein 5 |
| chr18_-_500692 | 0.14 |
ENST00000400256.5
|
COLEC12
|
collectin subfamily member 12 |
| chr14_-_21536884 | 0.14 |
ENST00000546363.5
|
SALL2
|
spalt like transcription factor 2 |
| chr6_-_46491431 | 0.14 |
ENST00000371374.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr21_+_46001300 | 0.14 |
ENST00000612273.2
ENST00000682634.1 |
COL6A1
|
collagen type VI alpha 1 chain |
| chr6_-_33746848 | 0.14 |
ENST00000634274.1
ENST00000293756.5 ENST00000451316.6 |
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr6_-_88166339 | 0.14 |
ENST00000369501.3
ENST00000551417.2 |
CNR1
|
cannabinoid receptor 1 |
| chr20_+_36214373 | 0.14 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr12_-_123972709 | 0.14 |
ENST00000545891.5
|
CCDC92
|
coiled-coil domain containing 92 |
| chr1_+_159171607 | 0.14 |
ENST00000368124.8
ENST00000368125.9 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr12_+_132687570 | 0.14 |
ENST00000317479.8
ENST00000543589.5 |
PXMP2
|
peroxisomal membrane protein 2 |
| chrX_+_150363306 | 0.14 |
ENST00000370401.7
ENST00000432680.7 |
MAMLD1
|
mastermind like domain containing 1 |
| chr14_+_20999255 | 0.14 |
ENST00000554422.5
ENST00000298681.5 |
SLC39A2
|
solute carrier family 39 member 2 |
| chrX_+_150363258 | 0.14 |
ENST00000683696.1
|
MAMLD1
|
mastermind like domain containing 1 |
| chr17_-_83051748 | 0.14 |
ENST00000320865.3
|
B3GNTL1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase like 1 |
| chr2_+_6917404 | 0.14 |
ENST00000320892.11
|
RNF144A
|
ring finger protein 144A |
| chr1_+_1001002 | 0.14 |
ENST00000624697.4
ENST00000624652.1 |
ISG15
|
ISG15 ubiquitin like modifier |
| chr17_-_82698637 | 0.14 |
ENST00000538809.6
ENST00000269347.10 ENST00000571995.6 |
RAB40B
|
RAB40B, member RAS oncogene family |
| chr9_-_21994345 | 0.14 |
ENST00000579755.2
ENST00000530628.2 |
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr16_+_176659 | 0.14 |
ENST00000320868.9
ENST00000397797.1 |
HBA1
|
hemoglobin subunit alpha 1 |
| chr8_+_96645221 | 0.13 |
ENST00000220763.10
|
CPQ
|
carboxypeptidase Q |
| chr3_-_9769910 | 0.13 |
ENST00000256460.8
|
CAMK1
|
calcium/calmodulin dependent protein kinase I |
| chr1_-_236065079 | 0.13 |
ENST00000264187.7
ENST00000366595.7 |
NID1
|
nidogen 1 |
| chr11_+_64234569 | 0.13 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr2_+_8682046 | 0.13 |
ENST00000331129.3
ENST00000396290.2 |
ID2
|
inhibitor of DNA binding 2 |
| chr1_-_156705575 | 0.13 |
ENST00000368222.8
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr4_+_4386498 | 0.13 |
ENST00000433139.6
|
NSG1
|
neuronal vesicle trafficking associated 1 |
| chr2_-_127106961 | 0.13 |
ENST00000376113.6
|
BIN1
|
bridging integrator 1 |
| chr11_+_308217 | 0.13 |
ENST00000602569.2
|
IFITM2
|
interferon induced transmembrane protein 2 |
| chr15_+_24675769 | 0.13 |
ENST00000329468.5
|
NPAP1
|
nuclear pore associated protein 1 |
| chr17_-_57955396 | 0.13 |
ENST00000577830.6
|
CUEDC1
|
CUE domain containing 1 |
| chr11_-_105035113 | 0.13 |
ENST00000526568.5
ENST00000531166.5 ENST00000534497.5 ENST00000527979.5 ENST00000533400.6 ENST00000528974.1 ENST00000525825.5 ENST00000353247.9 ENST00000446369.5 ENST00000436863.7 |
CASP1
|
caspase 1 |
| chr19_+_49860070 | 0.13 |
ENST00000597730.1
|
PTOV1
|
PTOV1 extended AT-hook containing adaptor protein |
| chr1_-_79006680 | 0.13 |
ENST00000370742.4
ENST00000656841.1 |
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr3_-_66500973 | 0.13 |
ENST00000383703.3
ENST00000273261.8 |
LRIG1
|
leucine rich repeats and immunoglobulin like domains 1 |
| chr4_+_4386256 | 0.13 |
ENST00000397958.5
|
NSG1
|
neuronal vesicle trafficking associated 1 |
| chr9_+_87498491 | 0.13 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1 |
| chr11_+_117199363 | 0.13 |
ENST00000392951.9
ENST00000525531.5 ENST00000278968.10 |
TAGLN
|
transgelin |
| chr3_-_169869833 | 0.13 |
ENST00000523069.1
ENST00000264676.9 ENST00000316428.10 |
LRRC31
|
leucine rich repeat containing 31 |
| chr9_+_137423350 | 0.13 |
ENST00000341349.6
ENST00000683555.1 ENST00000392815.2 ENST00000683683.1 |
NOXA1
|
NADPH oxidase activator 1 |
| chr2_+_233060295 | 0.13 |
ENST00000445964.6
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr19_+_44905785 | 0.13 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr21_-_43659460 | 0.13 |
ENST00000443485.1
ENST00000291560.7 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr2_+_27078598 | 0.13 |
ENST00000380320.9
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr1_-_1358524 | 0.12 |
ENST00000445648.5
ENST00000309212.11 |
MXRA8
|
matrix remodeling associated 8 |
| chr19_-_4455292 | 0.12 |
ENST00000394765.7
ENST00000592515.1 |
UBXN6
|
UBX domain protein 6 |
| chr12_-_91179355 | 0.12 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr4_-_993376 | 0.12 |
ENST00000398520.6
ENST00000398516.3 |
SLC26A1
|
solute carrier family 26 member 1 |
| chrX_-_107775951 | 0.12 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr16_+_56961942 | 0.12 |
ENST00000200676.8
ENST00000566128.1 |
CETP
|
cholesteryl ester transfer protein |
| chr19_+_49474208 | 0.12 |
ENST00000597551.6
ENST00000596435.5 ENST00000204637.6 ENST00000600429.5 |
FLT3LG
|
fms related receptor tyrosine kinase 3 ligand |
| chr8_+_95133940 | 0.12 |
ENST00000519516.1
|
PLEKHF2
|
pleckstrin homology and FYVE domain containing 2 |
| chr2_+_27051637 | 0.12 |
ENST00000451003.5
ENST00000360131.5 ENST00000323064.12 |
AGBL5
|
ATP/GTP binding protein like 5 |
| chr21_+_45405117 | 0.12 |
ENST00000651438.1
|
COL18A1
|
collagen type XVIII alpha 1 chain |
| chr20_+_64063481 | 0.12 |
ENST00000415602.5
|
TCEA2
|
transcription elongation factor A2 |
| chr16_-_28539004 | 0.12 |
ENST00000395641.2
|
NUPR1
|
nuclear protein 1, transcriptional regulator |
| chr2_+_172084748 | 0.12 |
ENST00000469444.6
|
DLX1
|
distal-less homeobox 1 |
| chr1_-_156677400 | 0.12 |
ENST00000368223.4
|
NES
|
nestin |
| chr2_+_238138661 | 0.12 |
ENST00000409223.2
|
KLHL30
|
kelch like family member 30 |
| chr6_-_31729478 | 0.12 |
ENST00000436437.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr9_-_21305313 | 0.12 |
ENST00000610521.2
|
IFNA5
|
interferon alpha 5 |
| chrX_+_43656289 | 0.12 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr22_-_39152622 | 0.12 |
ENST00000216133.10
|
CBX7
|
chromobox 7 |
| chr21_+_45455514 | 0.12 |
ENST00000355480.10
ENST00000359759.8 |
COL18A1
|
collagen type XVIII alpha 1 chain |
| chr10_+_132332273 | 0.12 |
ENST00000368613.8
|
LRRC27
|
leucine rich repeat containing 27 |
| chr19_+_18008162 | 0.12 |
ENST00000593560.6
ENST00000222250.5 |
ARRDC2
|
arrestin domain containing 2 |
| chr2_+_127418420 | 0.12 |
ENST00000234071.8
ENST00000429925.5 ENST00000442644.5 |
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr9_-_137459296 | 0.12 |
ENST00000371472.6
ENST00000371475.9 ENST00000371473.7 ENST00000371474.7 ENST00000437259.5 ENST00000265663.12 |
NSMF
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr21_+_42653734 | 0.12 |
ENST00000335512.8
ENST00000328862.10 ENST00000335440.10 ENST00000380328.6 ENST00000398225.7 ENST00000398227.7 ENST00000398229.7 ENST00000398232.7 ENST00000398234.7 ENST00000398236.7 ENST00000349112.7 ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr8_-_27611325 | 0.12 |
ENST00000523500.5
|
CLU
|
clusterin |
| chr9_+_128922283 | 0.12 |
ENST00000421063.6
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr1_-_156705742 | 0.12 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr8_-_27611424 | 0.12 |
ENST00000405140.7
|
CLU
|
clusterin |
| chr4_-_141132568 | 0.12 |
ENST00000506101.2
|
RNF150
|
ring finger protein 150 |
| chr4_-_56681588 | 0.12 |
ENST00000554144.5
ENST00000381260.7 |
HOPX
|
HOP homeobox |
| chr6_-_31728877 | 0.12 |
ENST00000437288.5
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr7_-_27662836 | 0.12 |
ENST00000265395.7
|
HIBADH
|
3-hydroxyisobutyrate dehydrogenase |
| chr2_+_171522466 | 0.11 |
ENST00000321348.9
ENST00000375252.3 |
CYBRD1
|
cytochrome b reductase 1 |
| chr5_+_172641241 | 0.11 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr7_-_45921264 | 0.11 |
ENST00000613132.5
ENST00000381083.9 ENST00000381086.9 |
IGFBP3
|
insulin like growth factor binding protein 3 |
| chr7_-_38363476 | 0.11 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr20_+_38724478 | 0.11 |
ENST00000217420.2
|
SLC32A1
|
solute carrier family 32 member 1 |
| chr10_-_59709842 | 0.11 |
ENST00000395348.8
|
SLC16A9
|
solute carrier family 16 member 9 |
| chr19_+_46601237 | 0.11 |
ENST00000597743.5
|
CALM3
|
calmodulin 3 |
| chr17_+_19378476 | 0.11 |
ENST00000395604.8
ENST00000482850.1 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr11_+_289155 | 0.11 |
ENST00000409655.5
|
PGGHG
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr6_+_83853576 | 0.11 |
ENST00000369687.2
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr7_+_134565098 | 0.11 |
ENST00000652743.1
|
AKR1B15
|
aldo-keto reductase family 1 member B15 |
| chr2_+_219572304 | 0.11 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha |
| chr14_+_95876385 | 0.11 |
ENST00000504119.1
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA |
| chr1_+_27342014 | 0.11 |
ENST00000618673.4
ENST00000318074.9 ENST00000616558.5 |
SYTL1
|
synaptotagmin like 1 |
| chr11_+_77821125 | 0.11 |
ENST00000526415.5
ENST00000393427.6 ENST00000527134.5 ENST00000304716.12 ENST00000630098.2 |
AAMDC
|
adipogenesis associated Mth938 domain containing |
| chr10_-_103452384 | 0.11 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr14_+_105472930 | 0.11 |
ENST00000483017.7
|
CRIP2
|
cysteine rich protein 2 |
| chr17_-_31859207 | 0.11 |
ENST00000302362.11
|
COPRS
|
coordinator of PRMT5 and differentiation stimulator |
| chrX_-_100410264 | 0.11 |
ENST00000373034.8
|
PCDH19
|
protocadherin 19 |
| chr9_-_127877665 | 0.11 |
ENST00000644144.2
|
AK1
|
adenylate kinase 1 |
| chr17_+_6651745 | 0.11 |
ENST00000542475.3
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr1_-_1361777 | 0.11 |
ENST00000477278.3
|
MXRA8
|
matrix remodeling associated 8 |
| chr6_+_89081787 | 0.11 |
ENST00000354922.3
|
PNRC1
|
proline rich nuclear receptor coactivator 1 |
| chr2_+_171522227 | 0.11 |
ENST00000409484.5
|
CYBRD1
|
cytochrome b reductase 1 |
| chr19_+_11798514 | 0.11 |
ENST00000323169.10
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr6_-_166382972 | 0.11 |
ENST00000341756.10
ENST00000621630.1 ENST00000621685.4 |
MPC1
|
mitochondrial pyruvate carrier 1 |
| chr13_-_100588731 | 0.11 |
ENST00000471912.1
ENST00000683975.1 |
GGACT
|
gamma-glutamylamine cyclotransferase |
| chr5_+_10564064 | 0.11 |
ENST00000296657.7
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr3_-_58211212 | 0.11 |
ENST00000461914.7
|
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr20_+_36541484 | 0.11 |
ENST00000346786.2
|
MYL9
|
myosin light chain 9 |
| chr9_-_127874964 | 0.11 |
ENST00000373156.5
|
AK1
|
adenylate kinase 1 |
| chr1_-_19288764 | 0.11 |
ENST00000361640.5
|
AKR7A3
|
aldo-keto reductase family 7 member A3 |
| chr11_+_77821187 | 0.11 |
ENST00000525409.5
|
AAMDC
|
adipogenesis associated Mth938 domain containing |
| chr19_-_52690478 | 0.11 |
ENST00000598536.5
ENST00000594682.6 ENST00000601257.5 |
ZNF83
|
zinc finger protein 83 |
| chr7_-_137846860 | 0.11 |
ENST00000288490.9
ENST00000614521.2 ENST00000424189.6 ENST00000446122.5 |
DGKI
|
diacylglycerol kinase iota |
| chr11_+_65014103 | 0.11 |
ENST00000246747.9
ENST00000529384.5 ENST00000533729.1 |
ARL2
|
ADP ribosylation factor like GTPase 2 |
| chr1_-_79006773 | 0.11 |
ENST00000671209.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr4_-_1683991 | 0.11 |
ENST00000472884.6
ENST00000489363.5 ENST00000308132.10 |
FAM53A
|
family with sequence similarity 53 member A |
| chr3_-_196968822 | 0.11 |
ENST00000412723.6
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis class Z |
| chr11_+_313391 | 0.11 |
ENST00000680802.1
ENST00000528780.5 ENST00000681761.1 ENST00000681821.1 ENST00000328221.5 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr1_-_156248013 | 0.11 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr11_+_12377524 | 0.11 |
ENST00000334956.15
|
PARVA
|
parvin alpha |
| chr11_+_117203135 | 0.11 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr10_-_132331818 | 0.11 |
ENST00000368619.3
ENST00000456004.1 ENST00000368620.2 |
STK32C
|
serine/threonine kinase 32C |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.3 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.1 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 0.4 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.4 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.5 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.3 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.2 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.1 | 0.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.3 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.3 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.0 | 0.2 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.2 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0010932 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.0 | GO:1903004 | regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.0 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.0 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.0 | 0.1 | GO:0034414 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.5 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.4 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0005985 | sucrose metabolic process(GO:0005985) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.0 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 | 0.3 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0016488 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0039526 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.1 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.0 | 0.0 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.0 | GO:1990009 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.0 | GO:0071878 | negative regulation of adrenergic receptor signaling pathway(GO:0071878) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.2 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) negative regulation of mitotic cell cycle DNA replication(GO:1903464) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.1 | GO:0046078 | pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.0 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.0 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:2001271 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.0 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.0 | GO:0045554 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.0 | 0.1 | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining(GO:2001033) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.0 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.3 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.9 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:2000342 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.0 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 0.0 | 0.2 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.3 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.0 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.1 | 0.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0060987 | lipid tube(GO:0060987) |
| 0.1 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.0 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.0 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.3 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 0.1 | 0.3 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.0 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |