Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
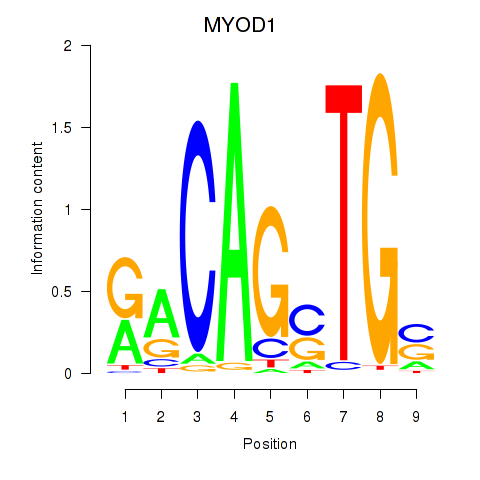
Results for MYOD1
Z-value: 0.48
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.4 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOD1 | hg38_v1_chr11_+_17719564_17719577 | -0.23 | 5.8e-01 | Click! |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41226648 | 0.38 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr17_-_41140487 | 0.34 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr6_+_150143018 | 0.19 |
ENST00000361131.5
|
PPP1R14C
|
protein phosphatase 1 regulatory inhibitor subunit 14C |
| chr11_-_111912871 | 0.19 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr17_+_41237998 | 0.19 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr21_+_29130630 | 0.18 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr19_+_35248375 | 0.18 |
ENST00000602122.5
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr11_+_1838970 | 0.17 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type |
| chr19_+_35248694 | 0.17 |
ENST00000361790.7
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35248656 | 0.17 |
ENST00000621372.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35248728 | 0.16 |
ENST00000602003.1
ENST00000360798.7 ENST00000354900.7 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr6_+_30884353 | 0.16 |
ENST00000428153.6
ENST00000376568.8 ENST00000452441.5 ENST00000515219.5 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_+_35248879 | 0.16 |
ENST00000347609.8
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr8_-_41665261 | 0.16 |
ENST00000522231.5
ENST00000314214.12 ENST00000348036.8 ENST00000522543.5 |
ANK1
|
ankyrin 1 |
| chr2_-_174764407 | 0.16 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr6_+_30884063 | 0.16 |
ENST00000511510.5
ENST00000376569.7 ENST00000376570.8 ENST00000504927.5 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_-_136550407 | 0.15 |
ENST00000354570.8
|
MAP7
|
microtubule associated protein 7 |
| chr14_+_75279637 | 0.15 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr7_-_76626127 | 0.15 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr17_-_41620801 | 0.14 |
ENST00000648859.1
|
KRT17
|
keratin 17 |
| chr12_-_6375556 | 0.13 |
ENST00000228916.7
|
SCNN1A
|
sodium channel epithelial 1 subunit alpha |
| chr19_+_44777860 | 0.13 |
ENST00000341505.4
ENST00000647358.2 |
CBLC
|
Cbl proto-oncogene C |
| chr2_+_30147516 | 0.12 |
ENST00000402708.5
|
YPEL5
|
yippee like 5 |
| chr11_-_95232514 | 0.12 |
ENST00000634898.1
ENST00000542176.1 ENST00000278499.6 |
SESN3
|
sestrin 3 |
| chr14_+_75280078 | 0.12 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr3_+_9933805 | 0.12 |
ENST00000684493.1
ENST00000673935.2 ENST00000684181.1 ENST00000683189.1 ENST00000383811.8 ENST00000452070.6 ENST00000682642.1 ENST00000684659.1 ENST00000491527.2 ENST00000326434.9 ENST00000682783.1 ENST00000683835.1 ENST00000682570.1 |
CRELD1
|
cysteine rich with EGF like domains 1 |
| chr5_+_66828762 | 0.11 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_1839452 | 0.11 |
ENST00000381906.5
|
TNNI2
|
troponin I2, fast skeletal type |
| chr6_+_30880780 | 0.11 |
ENST00000460944.6
ENST00000324771.12 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_+_119029711 | 0.11 |
ENST00000425699.3
|
NANOS1
|
nanos C2HC-type zinc finger 1 |
| chr11_+_1839602 | 0.11 |
ENST00000617947.4
ENST00000252898.11 |
TNNI2
|
troponin I2, fast skeletal type |
| chr11_+_57597563 | 0.11 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr11_+_44726811 | 0.11 |
ENST00000533202.5
ENST00000520358.7 ENST00000533080.5 ENST00000520999.6 |
TSPAN18
|
tetraspanin 18 |
| chr1_+_2073986 | 0.11 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr10_-_62816341 | 0.11 |
ENST00000242480.4
ENST00000637191.1 |
EGR2
|
early growth response 2 |
| chr20_-_32536383 | 0.11 |
ENST00000375678.7
|
NOL4L
|
nucleolar protein 4 like |
| chr11_-_123654939 | 0.11 |
ENST00000657191.1
|
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr2_+_64454145 | 0.10 |
ENST00000238875.10
|
LGALSL
|
galectin like |
| chr8_-_74321532 | 0.10 |
ENST00000342232.5
|
JPH1
|
junctophilin 1 |
| chr10_-_62816309 | 0.10 |
ENST00000411732.3
|
EGR2
|
early growth response 2 |
| chr11_+_57598184 | 0.10 |
ENST00000677625.1
ENST00000676670.1 |
SERPING1
|
serpin family G member 1 |
| chr19_+_35248994 | 0.10 |
ENST00000427250.5
ENST00000605618.6 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr14_+_67619911 | 0.10 |
ENST00000261783.4
|
ARG2
|
arginase 2 |
| chr19_-_38253238 | 0.10 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr20_+_59604527 | 0.10 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr6_-_136550819 | 0.10 |
ENST00000616617.4
ENST00000618822.4 |
MAP7
|
microtubule associated protein 7 |
| chr2_-_219571241 | 0.09 |
ENST00000373876.5
ENST00000603926.5 ENST00000373873.8 ENST00000289656.3 |
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr4_+_1793285 | 0.09 |
ENST00000440486.8
ENST00000412135.7 ENST00000481110.7 ENST00000340107.8 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr4_+_3463300 | 0.09 |
ENST00000340083.6
ENST00000507039.5 ENST00000643608.1 |
DOK7
|
docking protein 7 |
| chr16_-_31202733 | 0.09 |
ENST00000350605.4
ENST00000247470.10 |
PYCARD
|
PYD and CARD domain containing |
| chr4_+_1793776 | 0.09 |
ENST00000352904.6
|
FGFR3
|
fibroblast growth factor receptor 3 |
| chr1_-_154956086 | 0.09 |
ENST00000368463.8
ENST00000368460.7 ENST00000368465.5 |
PBXIP1
|
PBX homeobox interacting protein 1 |
| chr2_+_94871419 | 0.09 |
ENST00000295201.5
|
TEKT4
|
tektin 4 |
| chr14_-_24081928 | 0.09 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr2_+_232697362 | 0.09 |
ENST00000482666.5
ENST00000483164.5 ENST00000490229.5 ENST00000464805.5 ENST00000489328.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr12_+_66189178 | 0.08 |
ENST00000545837.1
|
IRAK3
|
interleukin 1 receptor associated kinase 3 |
| chr20_+_46894824 | 0.08 |
ENST00000327619.10
ENST00000497062.6 |
EYA2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr6_+_17281341 | 0.08 |
ENST00000379052.10
|
RBM24
|
RNA binding motif protein 24 |
| chr19_+_40717091 | 0.08 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr16_+_30375820 | 0.08 |
ENST00000566955.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr8_-_65789084 | 0.08 |
ENST00000379419.8
|
PDE7A
|
phosphodiesterase 7A |
| chr17_+_41105332 | 0.08 |
ENST00000391415.1
ENST00000617453.1 |
KRTAP4-9
|
keratin associated protein 4-9 |
| chr14_-_24081986 | 0.08 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr1_-_193186599 | 0.08 |
ENST00000367434.5
|
B3GALT2
|
beta-1,3-galactosyltransferase 2 |
| chr8_-_71356653 | 0.08 |
ENST00000388742.8
ENST00000388740.4 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_+_54123452 | 0.08 |
ENST00000620722.4
ENST00000490478.5 |
CACNA2D3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3 |
| chr6_-_24358036 | 0.08 |
ENST00000378454.8
|
DCDC2
|
doublecortin domain containing 2 |
| chr2_+_64453969 | 0.07 |
ENST00000464281.5
|
LGALSL
|
galectin like |
| chr17_-_55722857 | 0.07 |
ENST00000424486.3
|
TMEM100
|
transmembrane protein 100 |
| chr5_-_151224069 | 0.07 |
ENST00000355417.7
|
CCDC69
|
coiled-coil domain containing 69 |
| chr3_+_159852933 | 0.07 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr14_-_77141777 | 0.07 |
ENST00000319374.4
|
ZDHHC22
|
zinc finger DHHC-type palmitoyltransferase 22 |
| chr19_-_50823778 | 0.07 |
ENST00000301420.3
|
KLK1
|
kallikrein 1 |
| chr20_+_46894581 | 0.07 |
ENST00000357410.7
ENST00000611592.4 |
EYA2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr22_+_44702186 | 0.07 |
ENST00000336985.11
ENST00000403696.5 ENST00000457960.5 ENST00000361473.9 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 PRR5-ARHGAP8 readthrough |
| chr2_-_219571529 | 0.07 |
ENST00000404537.6
|
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr8_-_71356511 | 0.07 |
ENST00000419131.6
ENST00000388743.6 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr16_-_636253 | 0.07 |
ENST00000565163.5
|
METTL26
|
methyltransferase like 26 |
| chr17_+_60677822 | 0.07 |
ENST00000407086.8
ENST00000589222.5 ENST00000626960.2 ENST00000390652.9 |
BCAS3
|
BCAS3 microtubule associated cell migration factor |
| chr4_+_88378733 | 0.07 |
ENST00000273960.7
ENST00000380265.9 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_+_27234612 | 0.07 |
ENST00000319394.8
ENST00000361771.7 |
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr2_+_39665902 | 0.07 |
ENST00000281961.3
ENST00000618232.1 |
TMEM178A
|
transmembrane protein 178A |
| chr1_-_54406385 | 0.07 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr20_+_36573589 | 0.07 |
ENST00000373872.9
ENST00000650844.1 |
TGIF2
|
TGFB induced factor homeobox 2 |
| chr3_-_57079287 | 0.07 |
ENST00000338458.8
ENST00000468727.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr11_-_123654581 | 0.07 |
ENST00000392770.6
ENST00000530277.5 ENST00000299333.8 |
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr4_+_88378842 | 0.07 |
ENST00000264346.12
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_+_17205119 | 0.06 |
ENST00000375471.5
|
PADI1
|
peptidyl arginine deiminase 1 |
| chr8_+_119208322 | 0.06 |
ENST00000614891.5
|
MAL2
|
mal, T cell differentiation protein 2 |
| chr11_-_119381629 | 0.06 |
ENST00000260187.7
ENST00000455332.6 |
USP2
|
ubiquitin specific peptidase 2 |
| chr17_-_76570544 | 0.06 |
ENST00000640006.1
|
ENSG00000284526.1
|
novel protein |
| chr1_-_207032749 | 0.06 |
ENST00000359470.6
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr17_-_29622893 | 0.06 |
ENST00000345068.9
ENST00000584602.1 ENST00000388767.8 ENST00000580212.6 |
CORO6
|
coronin 6 |
| chr1_-_39901996 | 0.06 |
ENST00000397332.2
|
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr7_-_28958321 | 0.06 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats |
| chr17_+_75721451 | 0.06 |
ENST00000200181.8
|
ITGB4
|
integrin subunit beta 4 |
| chr10_-_123008784 | 0.06 |
ENST00000368886.10
|
IKZF5
|
IKAROS family zinc finger 5 |
| chr12_+_56079843 | 0.06 |
ENST00000549282.5
ENST00000549061.5 ENST00000683059.1 |
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr2_+_190137760 | 0.06 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr4_-_41214602 | 0.06 |
ENST00000508676.5
ENST00000506352.5 ENST00000295974.12 |
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr6_+_36130484 | 0.06 |
ENST00000373766.9
ENST00000211287.9 |
MAPK13
|
mitogen-activated protein kinase 13 |
| chr16_-_68372258 | 0.06 |
ENST00000568373.5
ENST00000563226.1 |
SMPD3
|
sphingomyelin phosphodiesterase 3 |
| chr3_-_100114488 | 0.06 |
ENST00000477258.2
ENST00000354552.7 ENST00000331335.9 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1 like |
| chr6_-_24911029 | 0.06 |
ENST00000259698.9
ENST00000644621.1 ENST00000644411.1 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr17_+_75721471 | 0.06 |
ENST00000450894.7
|
ITGB4
|
integrin subunit beta 4 |
| chr3_-_49813880 | 0.06 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7 |
| chr16_-_636270 | 0.06 |
ENST00000397665.6
ENST00000397666.6 ENST00000301686.13 ENST00000338401.8 ENST00000397664.8 ENST00000568830.1 ENST00000614890.4 |
METTL26
|
methyltransferase like 26 |
| chr16_+_85613252 | 0.06 |
ENST00000253458.12
ENST00000393243.5 |
GSE1
|
Gse1 coiled-coil protein |
| chr11_+_130159773 | 0.06 |
ENST00000278742.6
|
ST14
|
ST14 transmembrane serine protease matriptase |
| chr1_-_162023632 | 0.06 |
ENST00000367940.2
|
OLFML2B
|
olfactomedin like 2B |
| chr20_-_22585451 | 0.06 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr4_-_41214450 | 0.06 |
ENST00000513140.5
|
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr7_-_8262668 | 0.06 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1 |
| chr3_+_111998739 | 0.06 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr10_-_123008687 | 0.06 |
ENST00000617859.4
|
IKZF5
|
IKAROS family zinc finger 5 |
| chr12_+_116559381 | 0.06 |
ENST00000556529.4
|
MAP1LC3B2
|
microtubule associated protein 1 light chain 3 beta 2 |
| chr17_+_35147807 | 0.06 |
ENST00000394570.7
ENST00000268876.9 |
UNC45B
|
unc-45 myosin chaperone B |
| chr1_+_25616780 | 0.06 |
ENST00000374332.9
|
MAN1C1
|
mannosidase alpha class 1C member 1 |
| chr15_+_81196871 | 0.06 |
ENST00000559383.5
ENST00000394660.6 ENST00000683961.1 |
IL16
|
interleukin 16 |
| chr5_+_176810552 | 0.06 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A |
| chr4_-_41214535 | 0.05 |
ENST00000508593.6
|
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr7_+_18496269 | 0.05 |
ENST00000432645.6
|
HDAC9
|
histone deacetylase 9 |
| chr5_-_177509843 | 0.05 |
ENST00000510380.5
ENST00000357198.9 |
DOK3
|
docking protein 3 |
| chr12_+_68809002 | 0.05 |
ENST00000539479.6
ENST00000393415.7 ENST00000523991.5 ENST00000543323.5 ENST00000393416.7 |
MDM2
|
MDM2 proto-oncogene |
| chr6_-_24910695 | 0.05 |
ENST00000643623.1
ENST00000538035.6 ENST00000647136.1 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr1_+_206507520 | 0.05 |
ENST00000579436.7
|
RASSF5
|
Ras association domain family member 5 |
| chr17_-_58415628 | 0.05 |
ENST00000583753.5
|
RNF43
|
ring finger protein 43 |
| chr19_-_15479469 | 0.05 |
ENST00000292609.8
ENST00000340880.5 |
PGLYRP2
|
peptidoglycan recognition protein 2 |
| chr2_+_190649062 | 0.05 |
ENST00000409581.5
ENST00000337386.10 |
NAB1
|
NGFI-A binding protein 1 |
| chr19_-_7926106 | 0.05 |
ENST00000318978.6
|
CTXN1
|
cortexin 1 |
| chr8_-_41665200 | 0.05 |
ENST00000335651.6
|
ANK1
|
ankyrin 1 |
| chrX_-_129654946 | 0.05 |
ENST00000429967.3
|
APLN
|
apelin |
| chr10_-_101588126 | 0.05 |
ENST00000339310.7
ENST00000299206.8 ENST00000413344.5 ENST00000429502.1 ENST00000430045.1 ENST00000370172.5 ENST00000370162.8 ENST00000628479.2 |
POLL
|
DNA polymerase lambda |
| chr11_+_10455292 | 0.05 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr18_+_36297661 | 0.05 |
ENST00000257209.8
ENST00000590592.5 ENST00000359247.8 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr20_-_52191697 | 0.05 |
ENST00000361387.6
|
ZFP64
|
ZFP64 zinc finger protein |
| chr1_+_109712272 | 0.05 |
ENST00000369812.6
|
GSTM5
|
glutathione S-transferase mu 5 |
| chr1_+_167936559 | 0.05 |
ENST00000432587.6
ENST00000367843.7 ENST00000312263.10 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr11_+_64237420 | 0.05 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_+_206507546 | 0.05 |
ENST00000580449.5
ENST00000581503.6 |
RASSF5
|
Ras association domain family member 5 |
| chrX_-_65034707 | 0.05 |
ENST00000337990.2
|
ZC4H2
|
zinc finger C4H2-type containing |
| chr19_-_18919348 | 0.05 |
ENST00000349893.8
ENST00000351079.8 ENST00000600932.5 ENST00000262812.9 |
COPE
|
COPI coat complex subunit epsilon |
| chr1_-_150876571 | 0.05 |
ENST00000354396.6
ENST00000358595.10 ENST00000505755.5 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr2_-_27071628 | 0.05 |
ENST00000447619.5
ENST00000429985.1 ENST00000456793.2 |
OST4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr8_-_90645512 | 0.05 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr16_+_30372291 | 0.05 |
ENST00000568749.5
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_+_100612522 | 0.05 |
ENST00000393950.7
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr3_+_52420955 | 0.05 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr3_-_71725365 | 0.05 |
ENST00000425534.8
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr13_+_27620913 | 0.05 |
ENST00000637180.1
|
POLR1D
|
RNA polymerase I and III subunit D |
| chr20_+_44355692 | 0.05 |
ENST00000316673.8
ENST00000609795.5 ENST00000457232.5 ENST00000609262.5 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr7_+_100612430 | 0.05 |
ENST00000379527.6
|
MOSPD3
|
motile sperm domain containing 3 |
| chr2_-_156342348 | 0.05 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr9_-_98796494 | 0.05 |
ENST00000353234.5
|
ANKS6
|
ankyrin repeat and sterile alpha motif domain containing 6 |
| chr17_-_79952007 | 0.05 |
ENST00000574241.6
|
TBC1D16
|
TBC1 domain family member 16 |
| chr9_+_89605004 | 0.05 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr7_-_150978284 | 0.05 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2 |
| chr3_-_139539577 | 0.04 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1 |
| chr12_+_55932028 | 0.04 |
ENST00000394147.5
ENST00000551156.5 ENST00000553783.5 ENST00000557080.5 ENST00000432422.7 ENST00000556001.5 |
DGKA
|
diacylglycerol kinase alpha |
| chr17_+_75721327 | 0.04 |
ENST00000579662.5
|
ITGB4
|
integrin subunit beta 4 |
| chr3_+_50155024 | 0.04 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr21_+_25639251 | 0.04 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2 |
| chr10_-_124744280 | 0.04 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B |
| chr6_-_42142604 | 0.04 |
ENST00000356542.5
ENST00000341865.9 |
C6orf132
|
chromosome 6 open reading frame 132 |
| chr17_-_41836193 | 0.04 |
ENST00000435506.7
ENST00000415460.5 |
NT5C3B
|
5'-nucleotidase, cytosolic IIIB |
| chr14_+_104801082 | 0.04 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr3_-_186362223 | 0.04 |
ENST00000265022.8
|
DGKG
|
diacylglycerol kinase gamma |
| chr20_-_45827297 | 0.04 |
ENST00000372555.8
|
TNNC2
|
troponin C2, fast skeletal type |
| chr1_+_53062052 | 0.04 |
ENST00000395871.7
ENST00000673702.1 ENST00000673956.1 ENST00000312553.10 ENST00000371500.8 ENST00000618387.1 |
PODN
|
podocan |
| chr12_-_108827384 | 0.04 |
ENST00000326470.9
|
SSH1
|
slingshot protein phosphatase 1 |
| chr6_+_132570322 | 0.04 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr16_+_29806078 | 0.04 |
ENST00000545521.5
|
MAZ
|
MYC associated zinc finger protein |
| chr16_-_81077078 | 0.04 |
ENST00000565253.1
ENST00000378611.8 ENST00000299578.10 |
C16orf46
|
chromosome 16 open reading frame 46 |
| chr11_+_18602969 | 0.04 |
ENST00000636011.1
ENST00000542172.1 |
SPTY2D1OS
|
SPTY2D1 opposite strand |
| chr3_-_52452828 | 0.04 |
ENST00000496590.1
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr14_-_75981986 | 0.04 |
ENST00000238682.8
|
TGFB3
|
transforming growth factor beta 3 |
| chr11_-_117876892 | 0.04 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr16_+_83899079 | 0.04 |
ENST00000262430.6
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr19_-_11481044 | 0.04 |
ENST00000359227.8
|
ELAVL3
|
ELAV like RNA binding protein 3 |
| chr1_+_228149922 | 0.04 |
ENST00000366714.3
|
GJC2
|
gap junction protein gamma 2 |
| chr9_+_133534807 | 0.04 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS like 2 |
| chr2_+_120346130 | 0.04 |
ENST00000295228.4
|
INHBB
|
inhibin subunit beta B |
| chr9_+_4679555 | 0.04 |
ENST00000381858.5
ENST00000381854.4 |
CDC37L1
|
cell division cycle 37 like 1 |
| chr17_+_68205453 | 0.04 |
ENST00000674770.2
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr4_+_61200318 | 0.04 |
ENST00000683033.1
|
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr21_-_32813679 | 0.04 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr3_-_142149515 | 0.04 |
ENST00000475734.5
ENST00000467072.5 ENST00000489671.6 |
TFDP2
|
transcription factor Dp-2 |
| chr11_-_1486763 | 0.04 |
ENST00000329957.7
|
MOB2
|
MOB kinase activator 2 |
| chr12_-_6375209 | 0.04 |
ENST00000360168.7
|
SCNN1A
|
sodium channel epithelial 1 subunit alpha |
| chr11_-_695215 | 0.04 |
ENST00000382409.4
|
DEAF1
|
DEAF1 transcription factor |
| chr5_-_36241875 | 0.04 |
ENST00000381937.9
|
NADK2
|
NAD kinase 2, mitochondrial |
| chr3_+_49021605 | 0.04 |
ENST00000451378.2
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr14_-_105987068 | 0.04 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr12_-_42238203 | 0.04 |
ENST00000327791.8
ENST00000534854.7 |
YAF2
|
YY1 associated factor 2 |
| chr1_+_155078829 | 0.04 |
ENST00000368408.4
|
EFNA3
|
ephrin A3 |
| chr1_+_27879638 | 0.04 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2 |
| chr14_+_60249191 | 0.04 |
ENST00000395076.9
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A |
| chr16_-_636230 | 0.04 |
ENST00000568773.1
|
METTL26
|
methyltransferase like 26 |
| chr11_-_117876719 | 0.04 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr17_+_41249687 | 0.04 |
ENST00000334109.3
|
KRTAP9-4
|
keratin associated protein 9-4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0010931 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.1 | GO:2000405 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of T cell migration(GO:2000405) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.0 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0034031 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.0 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.0 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |