Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
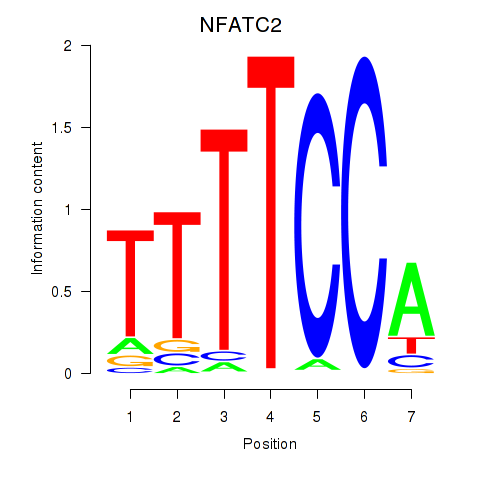
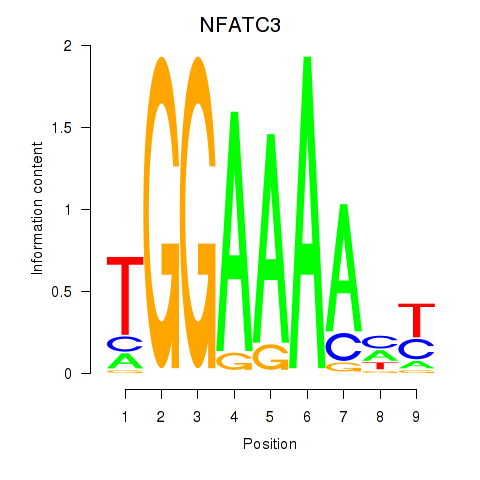
Results for NFATC2_NFATC3
Z-value: 0.56
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC2
|
ENSG00000101096.20 | nuclear factor of activated T cells 2 |
|
NFATC3
|
ENSG00000072736.19 | nuclear factor of activated T cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC2 | hg38_v1_chr20_-_51562829_51562853 | -0.70 | 5.1e-02 | Click! |
| NFATC3 | hg38_v1_chr16_+_68085861_68085968 | 0.34 | 4.1e-01 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_31447732 | 1.09 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr20_+_59604527 | 0.66 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_-_41703062 | 0.63 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr5_+_140875299 | 0.58 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chr6_-_136466858 | 0.54 |
ENST00000544465.5
|
MAP7
|
microtubule associated protein 7 |
| chr10_-_114684612 | 0.53 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_+_43935807 | 0.51 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr5_+_140841183 | 0.50 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr5_+_140827950 | 0.49 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr5_+_140848360 | 0.48 |
ENST00000532602.2
|
PCDHA9
|
protocadherin alpha 9 |
| chr1_+_46175079 | 0.46 |
ENST00000372003.6
|
TSPAN1
|
tetraspanin 1 |
| chr2_-_215138603 | 0.44 |
ENST00000272895.12
|
ABCA12
|
ATP binding cassette subfamily A member 12 |
| chr18_-_49849827 | 0.41 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr1_+_81800906 | 0.41 |
ENST00000674393.1
ENST00000674208.1 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr3_-_56775657 | 0.40 |
ENST00000413728.6
|
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr5_+_140882116 | 0.40 |
ENST00000289272.3
ENST00000409494.5 ENST00000617769.1 |
PCDHA13
|
protocadherin alpha 13 |
| chrX_+_106693838 | 0.39 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr5_+_140806929 | 0.39 |
ENST00000378125.4
ENST00000618834.1 ENST00000530339.2 ENST00000512229.6 ENST00000672575.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr5_+_66828762 | 0.38 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_126557691 | 0.38 |
ENST00000652209.1
|
LRATD2
|
LRAT domain containing 2 |
| chrX_-_15664798 | 0.38 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator |
| chr18_-_26549402 | 0.38 |
ENST00000408011.7
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr14_-_23154422 | 0.38 |
ENST00000422941.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr5_+_140868945 | 0.37 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11 |
| chr11_-_72781858 | 0.37 |
ENST00000537947.5
|
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr12_+_82686889 | 0.37 |
ENST00000321196.8
|
TMTC2
|
transmembrane O-mannosyltransferase targeting cadherins 2 |
| chr10_+_46375619 | 0.37 |
ENST00000584982.7
ENST00000613703.4 |
ANXA8L1
|
annexin A8 like 1 |
| chr11_-_72781833 | 0.35 |
ENST00000535054.1
ENST00000545082.5 |
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr14_+_64704380 | 0.34 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr18_+_63775395 | 0.32 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr6_+_106086316 | 0.31 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr20_+_46008900 | 0.31 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr18_-_26657401 | 0.31 |
ENST00000580191.5
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr5_+_168291599 | 0.31 |
ENST00000265293.9
|
WWC1
|
WW and C2 domain containing 1 |
| chr11_+_128693887 | 0.31 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_106098933 | 0.30 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr8_-_94262308 | 0.29 |
ENST00000297596.3
ENST00000396194.6 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr6_+_150143018 | 0.28 |
ENST00000361131.5
|
PPP1R14C
|
protein phosphatase 1 regulatory inhibitor subunit 14C |
| chr12_-_50222348 | 0.28 |
ENST00000552823.5
ENST00000552909.5 |
LIMA1
|
LIM domain and actin binding 1 |
| chr10_+_46375645 | 0.28 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1 |
| chr10_-_114684457 | 0.27 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr6_+_15246054 | 0.27 |
ENST00000341776.7
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr1_+_43933277 | 0.26 |
ENST00000414809.7
|
ARTN
|
artemin |
| chr8_-_15238423 | 0.26 |
ENST00000382080.6
|
SGCZ
|
sarcoglycan zeta |
| chr5_+_140834230 | 0.26 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr6_+_106087580 | 0.26 |
ENST00000424894.1
ENST00000648754.1 |
PRDM1
|
PR/SET domain 1 |
| chr13_-_40666600 | 0.26 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chr2_+_197705353 | 0.26 |
ENST00000282276.8
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr17_-_58415628 | 0.25 |
ENST00000583753.5
|
RNF43
|
ring finger protein 43 |
| chr1_-_207032749 | 0.24 |
ENST00000359470.6
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr12_-_52434363 | 0.24 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr12_-_6375556 | 0.24 |
ENST00000228916.7
|
SCNN1A
|
sodium channel epithelial 1 subunit alpha |
| chr19_-_55370455 | 0.24 |
ENST00000264563.7
ENST00000585513.1 ENST00000590625.5 |
IL11
|
interleukin 11 |
| chr15_+_43593054 | 0.23 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr2_-_112836702 | 0.23 |
ENST00000416750.1
ENST00000263341.7 ENST00000418817.5 |
IL1B
|
interleukin 1 beta |
| chr14_-_23154369 | 0.23 |
ENST00000453702.5
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr4_+_99816797 | 0.23 |
ENST00000512369.2
ENST00000296414.11 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr7_+_26291850 | 0.23 |
ENST00000338523.9
ENST00000446848.6 |
SNX10
|
sorting nexin 10 |
| chr5_-_79512794 | 0.22 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chr8_+_97887903 | 0.22 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr4_-_70666492 | 0.22 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr8_-_28386417 | 0.22 |
ENST00000521185.5
ENST00000520290.5 ENST00000344423.10 |
ZNF395
|
zinc finger protein 395 |
| chr8_-_28386073 | 0.21 |
ENST00000523095.5
ENST00000522795.1 |
ZNF395
|
zinc finger protein 395 |
| chr13_+_79481468 | 0.20 |
ENST00000620924.1
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr6_-_136550819 | 0.20 |
ENST00000616617.4
ENST00000618822.4 |
MAP7
|
microtubule associated protein 7 |
| chr5_+_140821598 | 0.20 |
ENST00000614258.1
ENST00000529859.2 ENST00000529619.5 |
PCDHA5
|
protocadherin alpha 5 |
| chr8_+_127736220 | 0.20 |
ENST00000524013.2
ENST00000621592.8 |
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr15_+_45430579 | 0.20 |
ENST00000558435.5
ENST00000344300.3 ENST00000396650.7 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr5_+_140786291 | 0.19 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1 |
| chr7_-_157010615 | 0.19 |
ENST00000252971.11
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr1_-_161021096 | 0.19 |
ENST00000537746.1
ENST00000368026.11 |
F11R
|
F11 receptor |
| chr1_-_205994439 | 0.19 |
ENST00000617991.4
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr9_-_123184233 | 0.19 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr18_+_22169580 | 0.18 |
ENST00000269216.10
|
GATA6
|
GATA binding protein 6 |
| chr6_-_11779606 | 0.18 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr13_-_85799400 | 0.17 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr5_+_140801028 | 0.17 |
ENST00000532566.3
ENST00000522353.3 |
PCDHA3
|
protocadherin alpha 3 |
| chr3_-_142029108 | 0.17 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr1_+_81800368 | 0.17 |
ENST00000674489.1
ENST00000674442.1 ENST00000674419.1 ENST00000674407.1 ENST00000674168.1 ENST00000674307.1 ENST00000674209.1 ENST00000370715.5 ENST00000370713.5 ENST00000319517.10 ENST00000627151.2 ENST00000370717.6 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr10_+_110207587 | 0.17 |
ENST00000332674.9
ENST00000453116.5 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr12_-_85836372 | 0.16 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chrX_-_134658450 | 0.16 |
ENST00000359237.9
|
PLAC1
|
placenta enriched 1 |
| chr3_+_111999326 | 0.16 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr6_-_170291053 | 0.16 |
ENST00000366756.4
|
DLL1
|
delta like canonical Notch ligand 1 |
| chr2_-_74440484 | 0.16 |
ENST00000305557.9
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr4_-_99435134 | 0.16 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_+_156147350 | 0.16 |
ENST00000435124.5
ENST00000633494.1 |
SEMA4A
|
semaphorin 4A |
| chr18_-_55422306 | 0.16 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr7_-_20217342 | 0.16 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr6_-_47042306 | 0.15 |
ENST00000371253.7
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr20_-_14337602 | 0.15 |
ENST00000378053.3
ENST00000341420.5 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr4_+_70334963 | 0.15 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr18_-_55422492 | 0.15 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr4_-_71784046 | 0.15 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr1_+_161721563 | 0.15 |
ENST00000367948.6
|
FCRLB
|
Fc receptor like B |
| chr17_+_76376581 | 0.15 |
ENST00000591651.5
ENST00000545180.5 |
SPHK1
|
sphingosine kinase 1 |
| chr15_-_52295792 | 0.14 |
ENST00000261839.12
|
MYO5C
|
myosin VC |
| chr11_+_34632464 | 0.14 |
ENST00000531794.5
|
EHF
|
ETS homologous factor |
| chr5_+_136059151 | 0.14 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr12_+_34022462 | 0.14 |
ENST00000538927.1
ENST00000266483.7 |
ALG10
|
ALG10 alpha-1,2-glucosyltransferase |
| chr13_+_79481446 | 0.14 |
ENST00000487865.5
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr8_-_17697654 | 0.13 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr1_-_183590439 | 0.13 |
ENST00000367535.8
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr12_-_70637405 | 0.13 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr2_+_86441341 | 0.13 |
ENST00000312912.10
ENST00000409064.5 |
KDM3A
|
lysine demethylase 3A |
| chr3_+_14402592 | 0.13 |
ENST00000622186.5
ENST00000621751.4 |
SLC6A6
|
solute carrier family 6 member 6 |
| chr19_+_40775511 | 0.13 |
ENST00000263369.4
ENST00000597140.5 |
MIA
|
MIA SH3 domain containing |
| chr1_+_24319511 | 0.13 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr3_+_141262614 | 0.13 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr18_+_58221535 | 0.13 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr12_-_54385727 | 0.13 |
ENST00000551109.5
ENST00000546970.5 |
ZNF385A
|
zinc finger protein 385A |
| chr20_+_56629296 | 0.13 |
ENST00000201031.3
|
TFAP2C
|
transcription factor AP-2 gamma |
| chr2_+_232697362 | 0.12 |
ENST00000482666.5
ENST00000483164.5 ENST00000490229.5 ENST00000464805.5 ENST00000489328.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr6_-_47042260 | 0.12 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr12_-_23951020 | 0.12 |
ENST00000441133.2
ENST00000545921.5 |
SOX5
|
SRY-box transcription factor 5 |
| chr3_-_125120813 | 0.12 |
ENST00000430155.6
|
SLC12A8
|
solute carrier family 12 member 8 |
| chr1_-_156705742 | 0.12 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr1_-_156705764 | 0.12 |
ENST00000621784.4
ENST00000368220.1 |
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr1_+_153776596 | 0.12 |
ENST00000458027.5
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr1_-_153565535 | 0.12 |
ENST00000368707.5
|
S100A2
|
S100 calcium binding protein A2 |
| chr17_+_40177460 | 0.12 |
ENST00000620260.6
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor like 1 |
| chr9_+_113150991 | 0.12 |
ENST00000259392.8
|
SLC31A2
|
solute carrier family 31 member 2 |
| chrX_-_117973579 | 0.11 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr5_-_78549151 | 0.11 |
ENST00000515007.6
|
LHFPL2
|
LHFPL tetraspan subfamily member 2 |
| chr1_+_101238090 | 0.11 |
ENST00000475289.2
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr1_+_213989691 | 0.11 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr9_+_100442271 | 0.11 |
ENST00000502978.1
|
MSANTD3-TMEFF1
|
MSANTD3-TMEFF1 readthrough |
| chr17_+_32487686 | 0.11 |
ENST00000584792.5
|
CDK5R1
|
cyclin dependent kinase 5 regulatory subunit 1 |
| chr18_-_55403682 | 0.11 |
ENST00000564228.5
ENST00000630828.2 |
TCF4
|
transcription factor 4 |
| chr13_+_97222296 | 0.11 |
ENST00000343600.8
ENST00000376673.7 ENST00000679496.1 ENST00000345429.10 |
MBNL2
|
muscleblind like splicing regulator 2 |
| chr17_-_48610971 | 0.11 |
ENST00000239165.9
|
HOXB7
|
homeobox B7 |
| chr14_+_75069577 | 0.11 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr5_+_136058849 | 0.11 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr1_+_24319342 | 0.11 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chrX_-_117973717 | 0.11 |
ENST00000262820.7
|
KLHL13
|
kelch like family member 13 |
| chr1_-_183590596 | 0.11 |
ENST00000418089.5
ENST00000413720.5 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr19_+_40775154 | 0.11 |
ENST00000594436.5
ENST00000597784.5 |
MIA
|
MIA SH3 domain containing |
| chrY_-_2787676 | 0.11 |
ENST00000383070.2
|
SRY
|
sex determining region Y |
| chr4_-_99435336 | 0.11 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_-_99016034 | 0.10 |
ENST00000448493.7
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr1_-_156705575 | 0.10 |
ENST00000368222.8
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr10_+_116545907 | 0.10 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr12_+_4269771 | 0.10 |
ENST00000676411.1
|
CCND2
|
cyclin D2 |
| chr1_+_247416149 | 0.10 |
ENST00000366497.6
ENST00000391828.8 |
NLRP3
|
NLR family pyrin domain containing 3 |
| chr4_+_68447453 | 0.10 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr17_+_36103819 | 0.10 |
ENST00000615863.2
ENST00000621626.1 |
CCL4
|
C-C motif chemokine ligand 4 |
| chr8_-_94436926 | 0.10 |
ENST00000481490.3
|
FSBP
|
fibrinogen silencer binding protein |
| chr3_+_14402576 | 0.10 |
ENST00000613060.4
|
SLC6A6
|
solute carrier family 6 member 6 |
| chr12_+_38316753 | 0.10 |
ENST00000551464.1
ENST00000308742.9 |
ALG10B
|
ALG10 alpha-1,2-glucosyltransferase B |
| chr2_+_102355750 | 0.10 |
ENST00000233957.7
|
IL18R1
|
interleukin 18 receptor 1 |
| chr10_+_119207560 | 0.10 |
ENST00000392870.3
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr18_-_70205655 | 0.10 |
ENST00000255674.11
ENST00000640769.2 |
RTTN
|
rotatin |
| chr1_+_247416166 | 0.10 |
ENST00000391827.3
ENST00000336119.8 |
NLRP3
|
NLR family pyrin domain containing 3 |
| chr7_+_148339452 | 0.10 |
ENST00000463592.3
|
CNTNAP2
|
contactin associated protein 2 |
| chr6_-_10415043 | 0.10 |
ENST00000379613.10
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr6_+_43771960 | 0.10 |
ENST00000230480.10
|
VEGFA
|
vascular endothelial growth factor A |
| chr17_+_16217198 | 0.10 |
ENST00000581006.5
ENST00000584797.5 ENST00000225609.10 ENST00000395844.8 ENST00000463810.2 |
PIGL
|
phosphatidylinositol glycan anchor biosynthesis class L |
| chr8_+_79611036 | 0.10 |
ENST00000220876.12
ENST00000518111.5 |
STMN2
|
stathmin 2 |
| chr9_+_122614738 | 0.10 |
ENST00000297913.3
|
OR1Q1
|
olfactory receptor family 1 subfamily Q member 1 |
| chr12_-_109309278 | 0.10 |
ENST00000299162.10
|
FOXN4
|
forkhead box N4 |
| chr14_+_75069632 | 0.10 |
ENST00000439583.2
ENST00000554763.2 ENST00000524913.3 ENST00000525046.2 ENST00000674086.1 ENST00000526130.2 ENST00000674094.1 ENST00000532198.2 |
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr18_+_79395942 | 0.10 |
ENST00000397790.6
|
NFATC1
|
nuclear factor of activated T cells 1 |
| chr1_+_174877430 | 0.10 |
ENST00000392064.6
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr2_+_102355881 | 0.10 |
ENST00000409599.5
|
IL18R1
|
interleukin 18 receptor 1 |
| chr14_-_21094488 | 0.09 |
ENST00000555270.5
|
ZNF219
|
zinc finger protein 219 |
| chr1_-_153549120 | 0.09 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr20_+_1894145 | 0.09 |
ENST00000400068.7
|
SIRPA
|
signal regulatory protein alpha |
| chr4_-_119322128 | 0.09 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr18_-_55402187 | 0.09 |
ENST00000630268.2
ENST00000570177.6 |
TCF4
|
transcription factor 4 |
| chr4_+_154743993 | 0.09 |
ENST00000336356.4
|
LRAT
|
lecithin retinol acyltransferase |
| chr14_-_23577457 | 0.09 |
ENST00000622501.4
|
JPH4
|
junctophilin 4 |
| chr11_-_57322197 | 0.09 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr12_-_109309218 | 0.09 |
ENST00000355216.5
|
FOXN4
|
forkhead box N4 |
| chr10_+_46375721 | 0.09 |
ENST00000616785.1
ENST00000611655.1 ENST00000619162.5 |
ENSG00000285402.1
ANXA8L1
|
novel transcript annexin A8 like 1 |
| chr6_-_27873525 | 0.09 |
ENST00000618305.2
|
H4C13
|
H4 clustered histone 13 |
| chrX_+_136648214 | 0.09 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr3_-_27722316 | 0.09 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr1_+_67166448 | 0.09 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor |
| chr10_+_69801874 | 0.09 |
ENST00000357811.8
|
COL13A1
|
collagen type XIII alpha 1 chain |
| chr8_-_90082871 | 0.09 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chrX_+_136648138 | 0.09 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr12_+_11649666 | 0.09 |
ENST00000396373.9
|
ETV6
|
ETS variant transcription factor 6 |
| chr18_-_55351977 | 0.09 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr17_-_7294592 | 0.09 |
ENST00000007699.10
|
YBX2
|
Y-box binding protein 2 |
| chr11_+_9461003 | 0.09 |
ENST00000438144.6
ENST00000396602.7 ENST00000526657.5 ENST00000299606.6 ENST00000534265.5 ENST00000412390.6 |
ZNF143
|
zinc finger protein 143 |
| chr4_-_142305826 | 0.09 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr20_+_1894462 | 0.08 |
ENST00000622179.4
|
SIRPA
|
signal regulatory protein alpha |
| chr17_-_40867200 | 0.08 |
ENST00000647902.1
ENST00000251643.5 |
KRT12
|
keratin 12 |
| chr1_-_146021724 | 0.08 |
ENST00000475797.1
ENST00000497365.5 ENST00000336751.11 ENST00000634927.1 ENST00000421822.2 |
HJV
|
hemojuvelin BMP co-receptor |
| chr8_-_85341705 | 0.08 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr17_+_29246852 | 0.08 |
ENST00000225387.8
|
CRYBA1
|
crystallin beta A1 |
| chr8_+_100158576 | 0.08 |
ENST00000388798.7
|
SPAG1
|
sperm associated antigen 1 |
| chr5_+_140855882 | 0.08 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr10_-_67696115 | 0.08 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr13_-_46142834 | 0.08 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr18_-_55589795 | 0.08 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chr6_-_134950081 | 0.08 |
ENST00000367847.2
ENST00000265605.7 ENST00000367845.6 |
ALDH8A1
|
aldehyde dehydrogenase 8 family member A1 |
| chr3_-_27722699 | 0.08 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr11_+_18396266 | 0.08 |
ENST00000540430.5
ENST00000379412.9 |
LDHA
|
lactate dehydrogenase A |
| chr1_-_153549238 | 0.08 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.6 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.8 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.2 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.3 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.2 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.1 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.7 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.3 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.3 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0036304 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.0 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.1 | GO:0060948 | coronary vein morphogenesis(GO:0003169) cardiac vascular smooth muscle cell development(GO:0060948) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.2 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 5.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:1904253 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:1902809 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:2000724 | mineralocorticoid receptor signaling pathway(GO:0031959) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.0 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0043696 | forebrain anterior/posterior pattern specification(GO:0021797) dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.0 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0071754 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.4 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 1.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.0 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |