Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
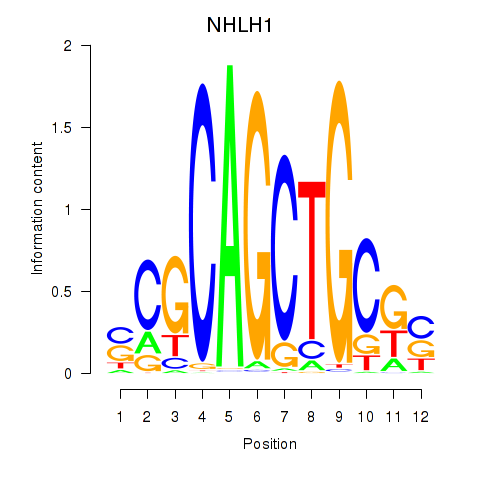
Results for NHLH1
Z-value: 0.85
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.6 | nescient helix-loop-helix 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg38_v1_chr1_+_160367061_160367078 | 0.14 | 7.4e-01 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_41124178 | 1.65 |
ENST00000394014.2
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr12_-_52493250 | 1.17 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr7_-_140062841 | 1.10 |
ENST00000263549.8
|
PARP12
|
poly(ADP-ribose) polymerase family member 12 |
| chr17_-_41118369 | 0.82 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr12_-_52473798 | 0.81 |
ENST00000252250.7
|
KRT6C
|
keratin 6C |
| chr16_+_22814154 | 0.79 |
ENST00000261374.4
|
HS3ST2
|
heparan sulfate-glucosamine 3-sulfotransferase 2 |
| chr17_-_41140487 | 0.75 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr20_+_59604527 | 0.68 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr5_+_53480619 | 0.66 |
ENST00000396947.7
ENST00000256759.8 |
FST
|
follistatin |
| chr6_+_30880780 | 0.65 |
ENST00000460944.6
ENST00000324771.12 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_81800906 | 0.64 |
ENST00000674393.1
ENST00000674208.1 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr6_+_150143018 | 0.59 |
ENST00000361131.5
|
PPP1R14C
|
protein phosphatase 1 regulatory inhibitor subunit 14C |
| chr5_+_167754918 | 0.57 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chr9_-_23821275 | 0.56 |
ENST00000380110.8
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr11_+_394196 | 0.53 |
ENST00000331563.7
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr9_-_27529705 | 0.52 |
ENST00000262244.6
|
MOB3B
|
MOB kinase activator 3B |
| chr8_-_124728273 | 0.49 |
ENST00000325064.9
ENST00000518547.6 |
MTSS1
|
MTSS I-BAR domain containing 1 |
| chr9_-_23826231 | 0.46 |
ENST00000397312.7
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr8_+_85463997 | 0.42 |
ENST00000285379.10
|
CA2
|
carbonic anhydrase 2 |
| chr18_+_11689210 | 0.41 |
ENST00000334049.11
|
GNAL
|
G protein subunit alpha L |
| chr6_+_125153649 | 0.41 |
ENST00000304877.17
ENST00000368402.9 ENST00000368388.6 ENST00000534000.6 |
TPD52L1
|
TPD52 like 1 |
| chr22_+_44702186 | 0.40 |
ENST00000336985.11
ENST00000403696.5 ENST00000457960.5 ENST00000361473.9 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 PRR5-ARHGAP8 readthrough |
| chr20_+_46008900 | 0.39 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr4_-_36244438 | 0.38 |
ENST00000303965.9
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_1882902 | 0.37 |
ENST00000231357.7
|
IRX4
|
iroquois homeobox 4 |
| chr17_+_41105332 | 0.36 |
ENST00000391415.1
ENST00000617453.1 |
KRTAP4-9
|
keratin associated protein 4-9 |
| chr6_+_125153793 | 0.35 |
ENST00000527711.5
|
TPD52L1
|
TPD52 like 1 |
| chr6_+_125154189 | 0.35 |
ENST00000532429.5
ENST00000534199.5 |
TPD52L1
|
TPD52 like 1 |
| chr17_+_41226648 | 0.34 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr8_+_24914942 | 0.34 |
ENST00000433454.3
|
NEFM
|
neurofilament medium |
| chr12_-_66678934 | 0.33 |
ENST00000545666.5
ENST00000398016.7 ENST00000359742.9 ENST00000538211.5 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr22_+_39994926 | 0.32 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F |
| chr1_-_37808168 | 0.31 |
ENST00000373044.3
|
YRDC
|
yrdC N6-threonylcarbamoyltransferase domain containing |
| chr19_+_45340760 | 0.31 |
ENST00000585434.5
|
KLC3
|
kinesin light chain 3 |
| chr2_+_68774782 | 0.31 |
ENST00000409030.7
ENST00000409220.5 |
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_+_13583762 | 0.31 |
ENST00000376057.8
ENST00000621990.5 ENST00000510906.5 |
PDPN
|
podoplanin |
| chr19_+_45340736 | 0.30 |
ENST00000391946.7
|
KLC3
|
kinesin light chain 3 |
| chr19_+_44777860 | 0.30 |
ENST00000341505.4
ENST00000647358.2 |
CBLC
|
Cbl proto-oncogene C |
| chr1_-_209806124 | 0.30 |
ENST00000367021.8
ENST00000542854.5 |
IRF6
|
interferon regulatory factor 6 |
| chr9_-_137054016 | 0.29 |
ENST00000312665.7
ENST00000355097.7 |
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr15_+_40252888 | 0.28 |
ENST00000559139.5
ENST00000560669.5 ENST00000542403.3 |
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr4_+_88378733 | 0.27 |
ENST00000273960.7
ENST00000380265.9 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr16_+_57639518 | 0.26 |
ENST00000540164.6
ENST00000568531.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr17_+_82735551 | 0.26 |
ENST00000300784.8
|
FN3K
|
fructosamine 3 kinase |
| chr17_-_41184895 | 0.26 |
ENST00000620667.1
ENST00000398472.2 |
KRTAP4-1
|
keratin associated protein 4-1 |
| chr14_-_53956811 | 0.26 |
ENST00000559087.5
ENST00000245451.9 |
BMP4
|
bone morphogenetic protein 4 |
| chr4_+_88378842 | 0.26 |
ENST00000264346.12
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr2_+_197705353 | 0.25 |
ENST00000282276.8
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr9_-_23826299 | 0.25 |
ENST00000380117.5
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr7_+_20330678 | 0.25 |
ENST00000537992.5
|
ITGB8
|
integrin subunit beta 8 |
| chr16_+_67431112 | 0.24 |
ENST00000326152.6
|
HSD11B2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr5_+_56815534 | 0.24 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr1_-_6602885 | 0.24 |
ENST00000377663.3
|
KLHL21
|
kelch like family member 21 |
| chr1_+_156284299 | 0.23 |
ENST00000456810.1
ENST00000405535.3 |
TMEM79
|
transmembrane protein 79 |
| chr4_-_138242325 | 0.23 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr19_+_45340774 | 0.23 |
ENST00000589837.5
|
KLC3
|
kinesin light chain 3 |
| chr2_-_234497035 | 0.23 |
ENST00000390645.2
ENST00000339728.6 |
ARL4C
|
ADP ribosylation factor like GTPase 4C |
| chr1_-_6602859 | 0.23 |
ENST00000377658.8
|
KLHL21
|
kelch like family member 21 |
| chr17_-_41168219 | 0.23 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr8_+_144148027 | 0.23 |
ENST00000423230.6
|
MROH1
|
maestro heat like repeat family member 1 |
| chr17_-_35795592 | 0.23 |
ENST00000615136.4
ENST00000605424.6 ENST00000612672.1 |
MMP28
|
matrix metallopeptidase 28 |
| chr15_+_40358207 | 0.23 |
ENST00000267889.5
|
DISP2
|
dispatched RND transporter family member 2 |
| chr22_-_37486357 | 0.22 |
ENST00000356998.8
ENST00000416983.7 ENST00000424765.2 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr18_+_58255433 | 0.22 |
ENST00000635997.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr20_-_14337602 | 0.22 |
ENST00000378053.3
ENST00000341420.5 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr4_-_80073057 | 0.21 |
ENST00000681710.1
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr20_+_63861498 | 0.21 |
ENST00000369916.5
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr2_+_195657179 | 0.21 |
ENST00000359634.10
|
SLC39A10
|
solute carrier family 39 member 10 |
| chr4_+_74445126 | 0.21 |
ENST00000395748.8
|
AREG
|
amphiregulin |
| chr7_+_129144691 | 0.20 |
ENST00000486685.3
|
TSPAN33
|
tetraspanin 33 |
| chr2_-_96145431 | 0.20 |
ENST00000288943.5
|
DUSP2
|
dual specificity phosphatase 2 |
| chr6_-_81752671 | 0.20 |
ENST00000320172.11
ENST00000369754.7 ENST00000369756.3 |
TENT5A
|
terminal nucleotidyltransferase 5A |
| chr22_+_44677044 | 0.20 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chr4_+_74445302 | 0.20 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr7_-_727611 | 0.19 |
ENST00000403562.5
|
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr17_+_57085092 | 0.19 |
ENST00000575322.1
ENST00000337714.8 |
AKAP1
|
A-kinase anchoring protein 1 |
| chr4_-_80073170 | 0.19 |
ENST00000403729.7
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr8_+_24913752 | 0.19 |
ENST00000518131.5
ENST00000221166.10 ENST00000437366.2 |
NEFM
|
neurofilament medium |
| chr5_+_69492767 | 0.18 |
ENST00000681041.1
ENST00000680098.1 ENST00000680784.1 ENST00000396442.7 ENST00000681895.1 |
OCLN
|
occludin |
| chr17_+_75516514 | 0.18 |
ENST00000333213.11
ENST00000545228.3 ENST00000680999.1 |
TSEN54
|
tRNA splicing endonuclease subunit 54 |
| chr12_+_41188301 | 0.18 |
ENST00000402685.7
|
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr1_+_32741779 | 0.18 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr8_-_130016395 | 0.18 |
ENST00000523509.5
|
CYRIB
|
CYFIP related Rac1 interactor B |
| chr8_-_130016414 | 0.18 |
ENST00000401979.6
ENST00000517654.5 ENST00000522361.1 ENST00000518167.5 |
CYRIB
|
CYFIP related Rac1 interactor B |
| chr22_-_24593038 | 0.18 |
ENST00000318753.13
|
LRRC75B
|
leucine rich repeat containing 75B |
| chrX_-_85379659 | 0.17 |
ENST00000262753.9
|
POF1B
|
POF1B actin binding protein |
| chr17_+_41237998 | 0.17 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr9_+_98807619 | 0.17 |
ENST00000375011.4
|
GALNT12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr6_-_30744537 | 0.17 |
ENST00000259874.6
ENST00000376377.2 |
IER3
|
immediate early response 3 |
| chr12_+_49623543 | 0.17 |
ENST00000548825.7
ENST00000261897.5 |
PRPF40B
|
pre-mRNA processing factor 40 homolog B |
| chr9_-_123268538 | 0.16 |
ENST00000360998.3
ENST00000348403.10 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr19_-_2051224 | 0.16 |
ENST00000309340.11
ENST00000589534.2 ENST00000250896.9 ENST00000589509.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr17_-_17206264 | 0.16 |
ENST00000321560.4
|
PLD6
|
phospholipase D family member 6 |
| chr16_+_2964216 | 0.16 |
ENST00000572045.5
ENST00000571007.5 ENST00000575885.5 ENST00000303746.10 ENST00000319500.10 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr1_+_156154371 | 0.16 |
ENST00000368282.1
|
SEMA4A
|
semaphorin 4A |
| chr14_+_23372809 | 0.16 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr8_+_97775775 | 0.16 |
ENST00000521545.7
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr6_+_18155399 | 0.16 |
ENST00000650836.2
ENST00000449850.2 ENST00000297792.9 |
KDM1B
|
lysine demethylase 1B |
| chr16_+_1706163 | 0.16 |
ENST00000250894.8
ENST00000673691.1 ENST00000356010.9 ENST00000610761.2 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr5_-_95284535 | 0.16 |
ENST00000515393.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr8_-_38150943 | 0.16 |
ENST00000276449.9
|
STAR
|
steroidogenic acute regulatory protein |
| chr8_+_144509049 | 0.16 |
ENST00000301327.5
|
MFSD3
|
major facilitator superfamily domain containing 3 |
| chr15_-_52295792 | 0.15 |
ENST00000261839.12
|
MYO5C
|
myosin VC |
| chr14_+_69879408 | 0.15 |
ENST00000361956.8
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr1_+_31413187 | 0.15 |
ENST00000373709.8
|
SERINC2
|
serine incorporator 2 |
| chr8_+_141128612 | 0.15 |
ENST00000518347.5
ENST00000262585.6 ENST00000520986.5 ENST00000523058.5 ENST00000518668.5 |
DENND3
|
DENN domain containing 3 |
| chr16_+_23835946 | 0.15 |
ENST00000321728.12
ENST00000643927.1 |
PRKCB
|
protein kinase C beta |
| chr14_+_63204859 | 0.15 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr8_+_141128581 | 0.15 |
ENST00000519811.6
|
DENND3
|
DENN domain containing 3 |
| chr16_+_2817230 | 0.15 |
ENST00000005995.8
ENST00000574813.5 |
PRSS21
|
serine protease 21 |
| chr1_+_14945775 | 0.15 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr18_+_46334197 | 0.15 |
ENST00000588679.1
ENST00000543885.2 |
RNF165
|
ring finger protein 165 |
| chr12_+_122021850 | 0.14 |
ENST00000261822.5
|
BCL7A
|
BAF chromatin remodeling complex subunit BCL7A |
| chr19_-_38253238 | 0.14 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr8_+_97775829 | 0.14 |
ENST00000517924.5
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr19_-_17448664 | 0.14 |
ENST00000341130.6
|
TMEM221
|
transmembrane protein 221 |
| chr1_+_202348727 | 0.14 |
ENST00000356764.6
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr17_-_32877106 | 0.14 |
ENST00000318217.10
ENST00000579584.5 ENST00000583621.1 |
MYO1D
|
myosin ID |
| chr12_+_7155867 | 0.14 |
ENST00000535313.2
ENST00000331148.5 |
CLSTN3
|
calsyntenin 3 |
| chr10_-_101588126 | 0.14 |
ENST00000339310.7
ENST00000299206.8 ENST00000413344.5 ENST00000429502.1 ENST00000430045.1 ENST00000370172.5 ENST00000370162.8 ENST00000628479.2 |
POLL
|
DNA polymerase lambda |
| chr12_+_6946468 | 0.14 |
ENST00000543115.5
ENST00000399448.5 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr3_-_45146351 | 0.14 |
ENST00000296129.6
ENST00000425231.2 |
CDCP1
|
CUB domain containing protein 1 |
| chr2_+_232697362 | 0.13 |
ENST00000482666.5
ENST00000483164.5 ENST00000490229.5 ENST00000464805.5 ENST00000489328.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr17_+_49575828 | 0.13 |
ENST00000328741.6
|
NXPH3
|
neurexophilin 3 |
| chr2_+_127645864 | 0.13 |
ENST00000544369.5
|
GPR17
|
G protein-coupled receptor 17 |
| chr4_-_80073465 | 0.13 |
ENST00000404191.5
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr6_+_14117764 | 0.13 |
ENST00000379153.4
|
CD83
|
CD83 molecule |
| chr14_+_32934383 | 0.13 |
ENST00000551634.6
|
NPAS3
|
neuronal PAS domain protein 3 |
| chr11_-_64166102 | 0.13 |
ENST00000255681.7
ENST00000675777.1 |
MACROD1
|
mono-ADP ribosylhydrolase 1 |
| chr16_+_11345429 | 0.13 |
ENST00000576027.1
ENST00000312499.6 ENST00000648619.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr16_+_74999312 | 0.13 |
ENST00000566250.5
ENST00000567962.5 |
ZNRF1
|
zinc and ring finger 1 |
| chr8_-_143815649 | 0.13 |
ENST00000356994.7
|
SCRIB
|
scribble planar cell polarity protein |
| chr12_+_109713817 | 0.12 |
ENST00000538780.2
|
FAM222A
|
family with sequence similarity 222 member A |
| chr2_+_48314637 | 0.12 |
ENST00000413569.5
ENST00000340553.8 |
FOXN2
|
forkhead box N2 |
| chr2_-_73284431 | 0.12 |
ENST00000521871.5
ENST00000520530.3 |
FBXO41
|
F-box protein 41 |
| chr8_+_127735597 | 0.12 |
ENST00000651626.1
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr16_-_29926155 | 0.12 |
ENST00000567795.3
ENST00000568000.6 ENST00000561540.2 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr14_+_70907661 | 0.12 |
ENST00000439984.7
|
PCNX1
|
pecanex 1 |
| chr18_-_55586092 | 0.12 |
ENST00000563888.6
ENST00000540999.5 ENST00000627685.2 |
TCF4
|
transcription factor 4 |
| chr3_-_165837412 | 0.12 |
ENST00000479451.5
ENST00000488954.1 ENST00000264381.8 |
BCHE
|
butyrylcholinesterase |
| chr11_+_44726811 | 0.12 |
ENST00000533202.5
ENST00000520358.7 ENST00000533080.5 ENST00000520999.6 |
TSPAN18
|
tetraspanin 18 |
| chr3_-_195909711 | 0.12 |
ENST00000333602.14
|
TNK2
|
tyrosine kinase non receptor 2 |
| chr15_+_50182215 | 0.12 |
ENST00000380902.8
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr16_+_27402167 | 0.12 |
ENST00000564089.5
ENST00000337929.8 |
IL21R
|
interleukin 21 receptor |
| chr11_-_66958366 | 0.12 |
ENST00000651036.1
ENST00000652125.1 ENST00000531614.6 ENST00000524491.6 ENST00000529047.6 ENST00000393960.7 ENST00000393958.7 ENST00000528403.6 ENST00000651854.1 |
PC
|
pyruvate carboxylase |
| chr2_-_72147819 | 0.12 |
ENST00000001146.7
ENST00000546307.5 ENST00000474509.1 |
CYP26B1
|
cytochrome P450 family 26 subfamily B member 1 |
| chr15_-_83284645 | 0.12 |
ENST00000345382.7
|
BNC1
|
basonuclin 1 |
| chr11_-_65606959 | 0.11 |
ENST00000532507.5
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr3_-_133895753 | 0.11 |
ENST00000460865.3
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr6_+_142301926 | 0.11 |
ENST00000296932.13
ENST00000367609.8 |
ADGRG6
|
adhesion G protein-coupled receptor G6 |
| chr17_-_28951443 | 0.11 |
ENST00000268756.7
ENST00000332830.9 ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr11_+_67303476 | 0.11 |
ENST00000376757.9
ENST00000308298.11 |
SSH3
|
slingshot protein phosphatase 3 |
| chr10_+_122163426 | 0.11 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr18_+_46334007 | 0.11 |
ENST00000269439.12
ENST00000590330.1 |
RNF165
|
ring finger protein 165 |
| chr9_-_115118145 | 0.11 |
ENST00000350763.9
|
TNC
|
tenascin C |
| chr9_-_115118198 | 0.11 |
ENST00000534839.1
ENST00000535648.5 |
TNC
|
tenascin C |
| chr2_+_195656734 | 0.11 |
ENST00000409086.7
|
SLC39A10
|
solute carrier family 39 member 10 |
| chr10_+_122163590 | 0.11 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr7_-_77416300 | 0.11 |
ENST00000257626.12
|
GSAP
|
gamma-secretase activating protein |
| chr12_+_119989869 | 0.11 |
ENST00000397558.6
|
BICDL1
|
BICD family like cargo adaptor 1 |
| chr2_+_232870284 | 0.11 |
ENST00000331342.4
|
SNORC
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr8_-_97277890 | 0.11 |
ENST00000322128.5
|
TSPYL5
|
TSPY like 5 |
| chr16_-_90019414 | 0.11 |
ENST00000002501.11
|
DBNDD1
|
dysbindin domain containing 1 |
| chr19_+_39196956 | 0.11 |
ENST00000339852.5
|
NCCRP1
|
NCCRP1, F-box associated domain containing |
| chr22_+_25742141 | 0.11 |
ENST00000536101.5
ENST00000335473.12 ENST00000407587.6 |
MYO18B
|
myosin XVIIIB |
| chr1_+_178513103 | 0.11 |
ENST00000319416.7
ENST00000367643.7 ENST00000367642.3 ENST00000367641.7 ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr20_+_50731571 | 0.11 |
ENST00000371610.7
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr7_+_151114597 | 0.11 |
ENST00000335367.7
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_2530256 | 0.10 |
ENST00000378453.4
|
HES5
|
hes family bHLH transcription factor 5 |
| chr10_+_122163672 | 0.10 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chrX_+_85244075 | 0.10 |
ENST00000276123.7
|
ZNF711
|
zinc finger protein 711 |
| chr10_+_69451456 | 0.10 |
ENST00000373290.7
|
TSPAN15
|
tetraspanin 15 |
| chr1_-_21622509 | 0.10 |
ENST00000374761.6
|
RAP1GAP
|
RAP1 GTPase activating protein |
| chr1_+_20589044 | 0.10 |
ENST00000375071.4
|
CDA
|
cytidine deaminase |
| chr7_+_20330893 | 0.10 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr8_+_103298836 | 0.10 |
ENST00000523739.5
ENST00000358755.5 |
FZD6
|
frizzled class receptor 6 |
| chr16_-_90019821 | 0.10 |
ENST00000568838.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr5_-_16936231 | 0.10 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr17_-_7393404 | 0.10 |
ENST00000575434.4
|
PLSCR3
|
phospholipid scramblase 3 |
| chr18_+_10454584 | 0.10 |
ENST00000355285.10
|
APCDD1
|
APC down-regulated 1 |
| chr19_+_7595830 | 0.10 |
ENST00000160298.9
ENST00000446248.4 |
CAMSAP3
|
calmodulin regulated spectrin associated protein family member 3 |
| chr14_-_24263162 | 0.10 |
ENST00000206765.11
ENST00000544573.5 |
TGM1
|
transglutaminase 1 |
| chr3_-_33659441 | 0.10 |
ENST00000650653.1
ENST00000480013.6 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr21_+_33070133 | 0.10 |
ENST00000382348.2
|
OLIG1
|
oligodendrocyte transcription factor 1 |
| chr13_-_36131286 | 0.10 |
ENST00000255448.8
ENST00000379892.4 |
DCLK1
|
doublecortin like kinase 1 |
| chr5_+_127290785 | 0.10 |
ENST00000503335.7
|
MEGF10
|
multiple EGF like domains 10 |
| chr1_-_3796478 | 0.10 |
ENST00000378251.3
|
LRRC47
|
leucine rich repeat containing 47 |
| chr17_+_7435416 | 0.09 |
ENST00000323206.2
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr3_-_197183806 | 0.09 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr11_+_7576975 | 0.09 |
ENST00000684215.1
ENST00000650027.1 |
PPFIBP2
|
PPFIA binding protein 2 |
| chr3_+_5187697 | 0.09 |
ENST00000256497.9
|
EDEM1
|
ER degradation enhancing alpha-mannosidase like protein 1 |
| chr14_+_30874497 | 0.09 |
ENST00000216361.9
|
COCH
|
cochlin |
| chr8_+_135457442 | 0.09 |
ENST00000355849.10
|
KHDRBS3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr7_-_139036017 | 0.09 |
ENST00000275766.2
|
ZC3HAV1L
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr5_+_76819022 | 0.09 |
ENST00000296677.5
|
F2RL1
|
F2R like trypsin receptor 1 |
| chr3_-_197183963 | 0.09 |
ENST00000653795.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr19_-_17334844 | 0.09 |
ENST00000159087.7
|
ANO8
|
anoctamin 8 |
| chr15_+_50182188 | 0.09 |
ENST00000267842.10
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr1_-_156677400 | 0.09 |
ENST00000368223.4
|
NES
|
nestin |
| chr10_-_124744280 | 0.09 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.5 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.4 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.3 | GO:2000007 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.2 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.2 | GO:0007308 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.1 | 0.2 | GO:0070859 | phthalate metabolic process(GO:0018963) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.4 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.4 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.7 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 0.1 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0043652 | skeletal muscle satellite cell activation(GO:0014719) engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.4 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 3.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.0 | GO:0034034 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 5.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |