Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
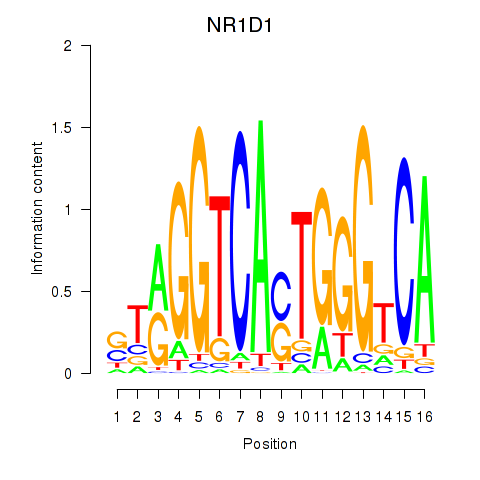
Results for NR1D1
Z-value: 0.16
Transcription factors associated with NR1D1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1D1
|
ENSG00000126368.6 | nuclear receptor subfamily 1 group D member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1D1 | hg38_v1_chr17_-_40100569_40100597 | 0.88 | 4.3e-03 | Click! |
Activity profile of NR1D1 motif
Sorted Z-values of NR1D1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_43933277 | 0.18 |
ENST00000414809.7
|
ARTN
|
artemin |
| chr1_+_200894892 | 0.13 |
ENST00000413687.3
|
INAVA
|
innate immunity activator |
| chr11_-_125496122 | 0.09 |
ENST00000527534.1
ENST00000278919.8 ENST00000366139.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 |
| chr8_+_104223320 | 0.09 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_-_6497096 | 0.08 |
ENST00000537245.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr8_+_104223344 | 0.06 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_20589044 | 0.06 |
ENST00000375071.4
|
CDA
|
cytidine deaminase |
| chr17_-_78132407 | 0.06 |
ENST00000322914.7
|
TMC6
|
transmembrane channel like 6 |
| chr17_-_39918589 | 0.06 |
ENST00000309481.11
|
GSDMB
|
gasdermin B |
| chr3_+_170037982 | 0.06 |
ENST00000355897.10
|
GPR160
|
G protein-coupled receptor 160 |
| chr17_-_39918606 | 0.06 |
ENST00000418519.6
ENST00000520542.5 |
GSDMB
|
gasdermin B |
| chr17_-_76532046 | 0.05 |
ENST00000590175.5
|
CYGB
|
cytoglobin |
| chr2_+_102736903 | 0.05 |
ENST00000639249.1
ENST00000454536.5 ENST00000409528.5 ENST00000409173.5 ENST00000488134.5 |
TMEM182
|
transmembrane protein 182 |
| chr17_+_41237998 | 0.05 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr10_-_102120246 | 0.05 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1 |
| chrX_-_49233309 | 0.04 |
ENST00000376251.5
ENST00000323022.10 ENST00000376265.2 |
CACNA1F
|
calcium voltage-gated channel subunit alpha1 F |
| chr3_-_185821092 | 0.04 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr1_-_11805924 | 0.04 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr19_-_49361475 | 0.04 |
ENST00000598810.5
|
TEAD2
|
TEA domain transcription factor 2 |
| chr11_+_60924452 | 0.04 |
ENST00000453848.7
ENST00000544065.5 ENST00000005286.8 |
TMEM132A
|
transmembrane protein 132A |
| chr16_+_3654683 | 0.04 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr9_+_98807619 | 0.03 |
ENST00000375011.4
|
GALNT12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr10_-_102120318 | 0.03 |
ENST00000673968.1
|
LDB1
|
LIM domain binding 1 |
| chr15_-_65611110 | 0.03 |
ENST00000567744.5
ENST00000568573.1 ENST00000562830.1 ENST00000569491.5 ENST00000561769.1 |
INTS14
|
integrator complex subunit 14 |
| chr15_-_65611236 | 0.03 |
ENST00000420799.7
ENST00000313182.6 ENST00000431261.6 ENST00000652388.1 ENST00000442903.3 |
INTS14
|
integrator complex subunit 14 |
| chr1_-_12618198 | 0.03 |
ENST00000616661.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr11_+_46719193 | 0.03 |
ENST00000311907.10
ENST00000530231.5 ENST00000442468.1 |
F2
|
coagulation factor II, thrombin |
| chr6_+_13574227 | 0.03 |
ENST00000680432.1
ENST00000681012.1 ENST00000397350.7 |
SIRT5
|
sirtuin 5 |
| chr6_+_31575557 | 0.03 |
ENST00000449264.3
|
TNF
|
tumor necrosis factor |
| chr8_+_100150621 | 0.03 |
ENST00000522439.1
ENST00000353107.8 |
POLR2K
|
RNA polymerase II, I and III subunit K |
| chr1_+_2467437 | 0.03 |
ENST00000449969.5
|
PLCH2
|
phospholipase C eta 2 |
| chr1_-_147773341 | 0.03 |
ENST00000430508.1
ENST00000621517.1 |
GJA5
|
gap junction protein alpha 5 |
| chr17_-_40100569 | 0.03 |
ENST00000246672.4
|
NR1D1
|
nuclear receptor subfamily 1 group D member 1 |
| chr2_+_112584434 | 0.03 |
ENST00000324913.9
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr17_-_2025289 | 0.03 |
ENST00000331238.7
|
RTN4RL1
|
reticulon 4 receptor like 1 |
| chr19_-_42423100 | 0.02 |
ENST00000597001.1
|
LIPE
|
lipase E, hormone sensitive type |
| chr2_+_112584586 | 0.02 |
ENST00000409719.1
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr19_-_14518383 | 0.02 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr17_-_58280928 | 0.02 |
ENST00000225275.4
|
MPO
|
myeloperoxidase |
| chr1_-_11805977 | 0.02 |
ENST00000376486.3
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr17_+_6996042 | 0.02 |
ENST00000251535.11
|
ALOX12
|
arachidonate 12-lipoxygenase, 12S type |
| chr19_-_4455292 | 0.02 |
ENST00000394765.7
ENST00000592515.1 |
UBXN6
|
UBX domain protein 6 |
| chr1_-_111449209 | 0.02 |
ENST00000235090.10
|
WDR77
|
WD repeat domain 77 |
| chr1_-_11805949 | 0.02 |
ENST00000376590.9
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr4_+_87608529 | 0.02 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein |
| chr2_-_102736819 | 0.02 |
ENST00000258436.10
|
MFSD9
|
major facilitator superfamily domain containing 9 |
| chr14_+_24070837 | 0.02 |
ENST00000537691.5
ENST00000397016.6 ENST00000560356.5 ENST00000558450.5 |
CPNE6
|
copine 6 |
| chr11_-_119381629 | 0.02 |
ENST00000260187.7
ENST00000455332.6 |
USP2
|
ubiquitin specific peptidase 2 |
| chr13_+_112969179 | 0.02 |
ENST00000535094.7
|
MCF2L
|
MCF.2 cell line derived transforming sequence like |
| chr10_+_22928010 | 0.02 |
ENST00000376528.8
|
ARMC3
|
armadillo repeat containing 3 |
| chr2_+_63589135 | 0.02 |
ENST00000432309.6
|
MDH1
|
malate dehydrogenase 1 |
| chr10_+_22928030 | 0.02 |
ENST00000409983.7
ENST00000298032.10 ENST00000409049.7 |
ARMC3
|
armadillo repeat containing 3 |
| chr22_-_37484505 | 0.02 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_+_98106852 | 0.02 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr22_+_37019735 | 0.01 |
ENST00000429360.6
ENST00000341116.7 ENST00000404393.5 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr2_+_63588953 | 0.01 |
ENST00000409908.5
ENST00000442225.5 ENST00000233114.13 ENST00000539945.7 ENST00000409476.5 ENST00000436321.5 |
MDH1
|
malate dehydrogenase 1 |
| chr2_+_74513441 | 0.01 |
ENST00000621092.1
|
TLX2
|
T cell leukemia homeobox 2 |
| chr8_-_109648825 | 0.01 |
ENST00000533895.5
ENST00000446070.6 ENST00000528331.5 ENST00000526302.5 ENST00000408908.6 ENST00000433638.1 ENST00000524720.5 |
SYBU
|
syntabulin |
| chr7_+_45000184 | 0.01 |
ENST00000544363.5
ENST00000541586.5 ENST00000258781.11 |
CCM2
|
CCM2 scaffold protein |
| chr1_+_197413827 | 0.01 |
ENST00000367397.1
ENST00000681519.1 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr10_+_133453936 | 0.01 |
ENST00000640237.1
|
SCART1
|
scavenger receptor family member expressed on T cells 1 |
| chr17_+_45844875 | 0.01 |
ENST00000329196.7
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr22_+_26621952 | 0.01 |
ENST00000354760.4
|
CRYBA4
|
crystallin beta A4 |
| chr7_+_94509793 | 0.01 |
ENST00000297273.9
|
CASD1
|
CAS1 domain containing 1 |
| chr3_-_182985926 | 0.01 |
ENST00000487822.5
ENST00000460412.6 ENST00000469954.5 |
DCUN1D1
|
defective in cullin neddylation 1 domain containing 1 |
| chr11_+_7513966 | 0.01 |
ENST00000299492.9
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr6_-_41747390 | 0.01 |
ENST00000356667.8
ENST00000373025.7 ENST00000425343.6 |
PGC
|
progastricsin |
| chr13_+_112968496 | 0.01 |
ENST00000397030.5
|
MCF2L
|
MCF.2 cell line derived transforming sequence like |
| chr2_+_126898908 | 0.01 |
ENST00000450035.5
|
TEX51
|
testis expressed 51 |
| chr2_+_126898876 | 0.01 |
ENST00000568484.6
ENST00000636457.1 |
TEX51
|
testis expressed 51 |
| chr9_-_37592564 | 0.01 |
ENST00000544379.1
ENST00000321301.7 ENST00000377773.9 ENST00000401811.3 |
TOMM5
|
translocase of outer mitochondrial membrane 5 |
| chr4_+_168921555 | 0.01 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_-_123271089 | 0.01 |
ENST00000535979.5
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr3_+_128794748 | 0.01 |
ENST00000674589.1
|
RAB7A
|
RAB7A, member RAS oncogene family |
| chr2_+_207711534 | 0.01 |
ENST00000392209.7
|
CCNYL1
|
cyclin Y like 1 |
| chr2_+_90114838 | 0.01 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr6_+_167111789 | 0.01 |
ENST00000400926.5
|
CCR6
|
C-C motif chemokine receptor 6 |
| chr9_-_33167296 | 0.01 |
ENST00000379731.5
ENST00000535206.5 |
B4GALT1
|
beta-1,4-galactosyltransferase 1 |
| chr1_+_203795614 | 0.00 |
ENST00000367210.3
ENST00000432282.5 ENST00000453771.5 ENST00000367214.5 ENST00000639812.1 ENST00000367212.7 ENST00000332127.8 ENST00000550078.2 |
ZC3H11A
ZBED6
|
zinc finger CCCH-type containing 11A zinc finger BED-type containing 6 |
| chr2_-_96505345 | 0.00 |
ENST00000310865.7
ENST00000451794.6 |
NEURL3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr3_-_48685835 | 0.00 |
ENST00000439518.5
ENST00000416649.6 ENST00000294129.7 |
NCKIPSD
|
NCK interacting protein with SH3 domain |
| chr14_-_91244669 | 0.00 |
ENST00000650645.1
|
GPR68
|
G protein-coupled receptor 68 |
| chr6_-_31970923 | 0.00 |
ENST00000495340.5
|
DXO
|
decapping exoribonuclease |
| chr19_+_21142058 | 0.00 |
ENST00000598331.1
|
ZNF431
|
zinc finger protein 431 |
| chr9_+_101028721 | 0.00 |
ENST00000374874.8
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr18_+_33578213 | 0.00 |
ENST00000681521.1
ENST00000269197.12 |
ASXL3
|
ASXL transcriptional regulator 3 |
| chr13_+_27270814 | 0.00 |
ENST00000241463.5
|
RASL11A
|
RAS like family 11 member A |
| chr7_-_129055158 | 0.00 |
ENST00000627585.2
|
TNPO3
|
transportin 3 |
| chr18_-_46757012 | 0.00 |
ENST00000315087.12
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr5_-_63962438 | 0.00 |
ENST00000323865.5
ENST00000506598.1 |
HTR1A
|
5-hydroxytryptamine receptor 1A |
| chr16_+_77191173 | 0.00 |
ENST00000248248.8
ENST00000439557.6 ENST00000545553.1 |
MON1B
|
MON1 homolog B, secretory trafficking associated |
| chr1_-_11805294 | 0.00 |
ENST00000413656.5
ENST00000376592.6 ENST00000376585.6 |
MTHFR
|
methylenetetrahydrofolate reductase |
| chr3_+_58237773 | 0.00 |
ENST00000478253.6
|
ABHD6
|
abhydrolase domain containing 6, acylglycerol lipase |
| chr19_+_21020675 | 0.00 |
ENST00000595401.1
|
ZNF430
|
zinc finger protein 430 |
| chr2_+_126898857 | 0.00 |
ENST00000643416.1
|
TEX51
|
testis expressed 51 |
| chr1_-_168543990 | 0.00 |
ENST00000367819.3
|
XCL2
|
X-C motif chemokine ligand 2 |
| chr2_+_88885397 | 0.00 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr7_-_129054869 | 0.00 |
ENST00000471166.1
|
TNPO3
|
transportin 3 |
| chr7_-_129055093 | 0.00 |
ENST00000482320.5
ENST00000265388.10 ENST00000471234.5 |
TNPO3
|
transportin 3 |
| chr10_+_88586762 | 0.00 |
ENST00000371939.7
|
LIPJ
|
lipase family member J |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1D1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |