Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
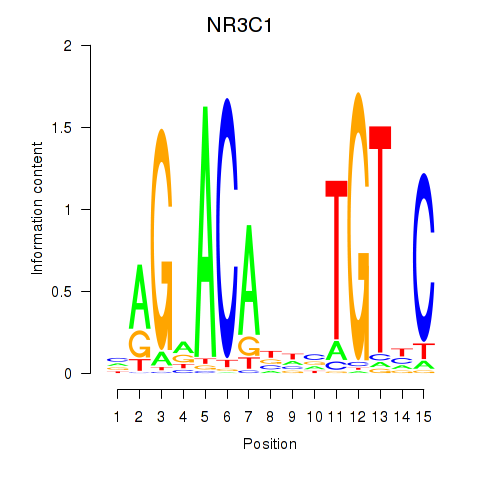
Results for NR3C1
Z-value: 0.75
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.15 | nuclear receptor subfamily 3 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg38_v1_chr5_-_143434677_143434713, hg38_v1_chr5_-_143404129_143404277 | -0.53 | 1.7e-01 | Click! |
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_151762899 | 0.47 |
ENST00000635322.1
ENST00000321531.10 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr3_-_190449782 | 0.39 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr15_-_21718245 | 0.36 |
ENST00000630556.1
|
ENSG00000281179.1
|
novel gene identicle to IGHV1OR15-1 |
| chr19_+_35140022 | 0.35 |
ENST00000588081.5
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_+_233060295 | 0.35 |
ENST00000445964.6
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr17_+_75979211 | 0.34 |
ENST00000397640.6
ENST00000588202.5 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 subunit of CST complex |
| chr16_+_56682461 | 0.33 |
ENST00000562939.1
ENST00000394485.5 ENST00000567563.1 |
MT1X
ENSG00000259827.1
|
metallothionein 1X novel transcript |
| chr20_+_64063481 | 0.33 |
ENST00000415602.5
|
TCEA2
|
transcription elongation factor A2 |
| chr3_-_183116075 | 0.33 |
ENST00000492597.5
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 |
| chr11_+_308408 | 0.33 |
ENST00000399815.2
|
ENSG00000288681.1
|
novel protein |
| chr7_+_76424922 | 0.32 |
ENST00000394857.8
|
ZP3
|
zona pellucida glycoprotein 3 |
| chr8_+_90001448 | 0.32 |
ENST00000519410.5
ENST00000522161.5 ENST00000220764.7 ENST00000517761.5 ENST00000520227.1 |
DECR1
|
2,4-dienoyl-CoA reductase 1 |
| chr16_+_56632651 | 0.29 |
ENST00000379818.4
ENST00000570233.1 |
MT1M
|
metallothionein 1M |
| chr3_-_119559529 | 0.28 |
ENST00000478182.5
|
CD80
|
CD80 molecule |
| chr11_-_106077401 | 0.27 |
ENST00000526793.5
|
KBTBD3
|
kelch repeat and BTB domain containing 3 |
| chr3_+_152300135 | 0.27 |
ENST00000465907.6
ENST00000492948.5 ENST00000485509.5 ENST00000464596.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr15_+_24675769 | 0.27 |
ENST00000329468.5
|
NPAP1
|
nuclear pore associated protein 1 |
| chr22_-_16592810 | 0.26 |
ENST00000359963.4
|
CCT8L2
|
chaperonin containing TCP1 subunit 8 like 2 |
| chr22_+_24607602 | 0.25 |
ENST00000447416.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr16_+_67173935 | 0.25 |
ENST00000566871.5
|
NOL3
|
nucleolar protein 3 |
| chr17_+_4715438 | 0.25 |
ENST00000571206.1
|
ARRB2
|
arrestin beta 2 |
| chr20_+_157447 | 0.25 |
ENST00000382388.4
|
DEFB127
|
defensin beta 127 |
| chr6_+_26045374 | 0.24 |
ENST00000612966.3
|
H3C3
|
H3 clustered histone 3 |
| chr17_+_68249200 | 0.24 |
ENST00000577985.5
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr7_+_150991005 | 0.23 |
ENST00000297494.8
|
NOS3
|
nitric oxide synthase 3 |
| chr6_+_18387326 | 0.23 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B |
| chr1_+_70411241 | 0.23 |
ENST00000370938.8
ENST00000346806.2 |
CTH
|
cystathionine gamma-lyase |
| chr16_-_48247533 | 0.23 |
ENST00000356608.7
ENST00000569991.1 |
ABCC11
|
ATP binding cassette subfamily C member 11 |
| chr11_-_106077313 | 0.23 |
ENST00000531837.2
ENST00000534815.1 |
KBTBD3
|
kelch repeat and BTB domain containing 3 |
| chr17_-_75765136 | 0.23 |
ENST00000592997.6
ENST00000588479.6 ENST00000225614.6 |
GALK1
|
galactokinase 1 |
| chr16_+_3065380 | 0.22 |
ENST00000551122.5
ENST00000548807.5 ENST00000528163.6 |
IL32
|
interleukin 32 |
| chr15_-_22160868 | 0.22 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr7_+_142332182 | 0.22 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr19_+_8413270 | 0.22 |
ENST00000381035.8
ENST00000595142.5 ENST00000601724.5 ENST00000601283.5 ENST00000215555.7 ENST00000595213.1 |
MARCHF2
|
membrane associated ring-CH-type finger 2 |
| chr15_-_89814845 | 0.22 |
ENST00000679248.1
ENST00000300060.7 ENST00000560137.2 |
ANPEP
|
alanyl aminopeptidase, membrane |
| chr13_+_87671354 | 0.21 |
ENST00000683689.1
|
SLITRK5
|
SLIT and NTRK like family member 5 |
| chr17_-_19386785 | 0.21 |
ENST00000497081.6
|
MFAP4
|
microfibril associated protein 4 |
| chr3_-_121660892 | 0.21 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr11_+_5689780 | 0.21 |
ENST00000379965.8
ENST00000454828.5 |
TRIM22
|
tripartite motif containing 22 |
| chr1_+_75796867 | 0.21 |
ENST00000263187.4
|
MSH4
|
mutS homolog 4 |
| chr7_+_150991087 | 0.20 |
ENST00000461406.5
|
NOS3
|
nitric oxide synthase 3 |
| chr2_+_233729042 | 0.20 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr1_+_220690354 | 0.20 |
ENST00000294889.6
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr17_+_42552920 | 0.20 |
ENST00000585807.6
ENST00000225929.5 |
HSD17B1
|
hydroxysteroid 17-beta dehydrogenase 1 |
| chr4_-_151227881 | 0.20 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr22_+_24607658 | 0.19 |
ENST00000451366.5
ENST00000428855.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr1_-_183653307 | 0.19 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr11_+_117200188 | 0.19 |
ENST00000529792.5
|
TAGLN
|
transgelin |
| chr17_+_77405070 | 0.19 |
ENST00000585930.5
|
SEPTIN9
|
septin 9 |
| chr17_+_7558296 | 0.19 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr10_+_5364955 | 0.19 |
ENST00000380433.5
|
UCN3
|
urocortin 3 |
| chr1_-_27490130 | 0.19 |
ENST00000618852.5
|
WASF2
|
WASP family member 2 |
| chr16_+_3065311 | 0.19 |
ENST00000534507.5
ENST00000613483.4 ENST00000531965.5 ENST00000396887.7 |
IL32
|
interleukin 32 |
| chr21_-_43076362 | 0.19 |
ENST00000359624.7
ENST00000352178.9 |
CBS
|
cystathionine beta-synthase |
| chr10_-_28282086 | 0.19 |
ENST00000375719.7
ENST00000375732.5 |
MPP7
|
membrane palmitoylated protein 7 |
| chr8_+_26390362 | 0.19 |
ENST00000518611.5
|
BNIP3L
|
BCL2 interacting protein 3 like |
| chr14_-_106737547 | 0.19 |
ENST00000632209.1
|
IGHV1-69-2
|
immunoglobulin heavy variable 1-69-2 |
| chr21_-_6468040 | 0.19 |
ENST00000618024.4
ENST00000617706.4 |
CBSL
|
cystathionine beta-synthase like |
| chr1_-_85404494 | 0.18 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr15_-_72197772 | 0.18 |
ENST00000309731.12
|
GRAMD2A
|
GRAM domain containing 2A |
| chr16_+_3065297 | 0.18 |
ENST00000325568.9
|
IL32
|
interleukin 32 |
| chr14_-_22957100 | 0.18 |
ENST00000555367.5
|
HAUS4
|
HAUS augmin like complex subunit 4 |
| chrX_-_2968236 | 0.18 |
ENST00000684117.1
ENST00000672761.1 ENST00000672027.1 ENST00000672606.1 ENST00000673032.1 ENST00000540563.6 |
ARSL
|
arylsulfatase L |
| chr14_-_22957128 | 0.18 |
ENST00000342454.12
ENST00000555986.5 ENST00000554516.5 ENST00000347758.6 ENST00000206474.11 ENST00000555040.5 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr4_-_687325 | 0.18 |
ENST00000503156.5
|
SLC49A3
|
solute carrier family 49 member 3 |
| chr21_-_44801769 | 0.18 |
ENST00000330942.9
|
UBE2G2
|
ubiquitin conjugating enzyme E2 G2 |
| chr6_-_37499852 | 0.18 |
ENST00000373408.4
|
CCDC167
|
coiled-coil domain containing 167 |
| chr11_-_2903490 | 0.18 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chr14_-_22957061 | 0.17 |
ENST00000557591.5
ENST00000541587.6 ENST00000490506.5 ENST00000554406.1 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr21_-_30372265 | 0.17 |
ENST00000399889.4
|
KRTAP13-2
|
keratin associated protein 13-2 |
| chr14_+_60249191 | 0.17 |
ENST00000395076.9
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A |
| chr7_+_40134966 | 0.17 |
ENST00000401647.7
ENST00000628514.3 ENST00000335693.9 ENST00000416370.2 |
SUGCT
|
succinyl-CoA:glutarate-CoA transferase |
| chr4_-_99352754 | 0.17 |
ENST00000639454.1
|
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr11_-_114400417 | 0.17 |
ENST00000325636.8
ENST00000623205.2 |
C11orf71
|
chromosome 11 open reading frame 71 |
| chr15_+_88635626 | 0.16 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr1_+_22653228 | 0.16 |
ENST00000509305.6
|
C1QB
|
complement C1q B chain |
| chr10_+_69088096 | 0.16 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr6_+_79631322 | 0.16 |
ENST00000369838.6
|
SH3BGRL2
|
SH3 domain binding glutamate rich protein like 2 |
| chr8_+_95133940 | 0.16 |
ENST00000519516.1
|
PLEKHF2
|
pleckstrin homology and FYVE domain containing 2 |
| chr12_-_49897056 | 0.16 |
ENST00000552863.5
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr1_-_67833448 | 0.16 |
ENST00000370982.4
|
GNG12
|
G protein subunit gamma 12 |
| chr1_+_207088825 | 0.16 |
ENST00000367078.8
|
C4BPB
|
complement component 4 binding protein beta |
| chr17_-_8124084 | 0.15 |
ENST00000317814.8
ENST00000577735.1 ENST00000541682.7 |
HES7
|
hes family bHLH transcription factor 7 |
| chr15_+_31366138 | 0.15 |
ENST00000558844.1
|
KLF13
|
Kruppel like factor 13 |
| chr15_-_45201094 | 0.15 |
ENST00000561278.1
ENST00000290894.12 |
SHF
|
Src homology 2 domain containing F |
| chr1_+_207089233 | 0.15 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr3_-_42875871 | 0.15 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chrX_-_115234088 | 0.15 |
ENST00000317135.13
|
LRCH2
|
leucine rich repeats and calponin homology domain containing 2 |
| chr21_-_32813679 | 0.15 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr2_+_190180447 | 0.15 |
ENST00000409870.1
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr1_+_207089283 | 0.15 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr9_-_37904085 | 0.15 |
ENST00000377716.6
ENST00000242275.7 |
SLC25A51
|
solute carrier family 25 member 51 |
| chr1_+_22653189 | 0.15 |
ENST00000432749.6
|
C1QB
|
complement C1q B chain |
| chr2_-_42764116 | 0.15 |
ENST00000378661.3
|
OXER1
|
oxoeicosanoid receptor 1 |
| chr15_+_81299416 | 0.15 |
ENST00000558332.3
|
IL16
|
interleukin 16 |
| chr1_-_36397880 | 0.14 |
ENST00000315732.3
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr16_+_4624811 | 0.14 |
ENST00000415496.5
ENST00000262370.12 ENST00000587747.5 ENST00000399577.9 ENST00000588994.5 ENST00000586183.5 |
MGRN1
|
mahogunin ring finger 1 |
| chr11_+_5449323 | 0.14 |
ENST00000641930.1
|
OR51I2
|
olfactory receptor family 51 subfamily I member 2 |
| chrX_-_115234042 | 0.14 |
ENST00000538422.2
|
LRCH2
|
leucine rich repeats and calponin homology domain containing 2 |
| chr17_-_74868616 | 0.14 |
ENST00000579893.1
ENST00000544854.5 |
FDXR
|
ferredoxin reductase |
| chr19_-_42412347 | 0.14 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase E, hormone sensitive type |
| chr1_-_182391323 | 0.14 |
ENST00000642379.1
|
GLUL
|
glutamate-ammonia ligase |
| chr6_-_90586883 | 0.14 |
ENST00000369325.7
ENST00000369327.7 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr10_-_75109085 | 0.14 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr5_-_142012973 | 0.14 |
ENST00000503794.5
ENST00000510194.5 ENST00000504424.1 ENST00000513454.5 ENST00000311337.11 ENST00000503229.5 ENST00000500692.6 ENST00000504139.5 ENST00000505689.5 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr10_-_75109172 | 0.14 |
ENST00000372700.7
ENST00000473072.2 ENST00000491677.6 ENST00000372702.7 |
DUSP13
|
dual specificity phosphatase 13 |
| chr1_-_1214146 | 0.14 |
ENST00000379236.4
|
TNFRSF4
|
TNF receptor superfamily member 4 |
| chr1_+_207089195 | 0.14 |
ENST00000452902.6
|
C4BPB
|
complement component 4 binding protein beta |
| chr1_+_165827574 | 0.14 |
ENST00000367879.9
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_+_145964675 | 0.14 |
ENST00000369314.2
ENST00000369313.7 |
POLR3GL
|
RNA polymerase III subunit GL |
| chr8_-_120445092 | 0.14 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr1_-_182391363 | 0.14 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase |
| chr19_+_15673069 | 0.14 |
ENST00000550308.6
ENST00000551607.5 |
CYP4F12
|
cytochrome P450 family 4 subfamily F member 12 |
| chr20_+_64063105 | 0.14 |
ENST00000395053.7
ENST00000343484.10 ENST00000339217.8 |
TCEA2
|
transcription elongation factor A2 |
| chr1_-_182391783 | 0.14 |
ENST00000331872.11
ENST00000339526.8 |
GLUL
|
glutamate-ammonia ligase |
| chr20_+_36573458 | 0.14 |
ENST00000373874.6
|
TGIF2
|
TGFB induced factor homeobox 2 |
| chr16_+_33009175 | 0.14 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr8_-_120445140 | 0.14 |
ENST00000306185.8
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr18_+_58862904 | 0.14 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532 |
| chr6_-_55579160 | 0.14 |
ENST00000370850.6
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase like 1 |
| chr1_+_234373439 | 0.14 |
ENST00000366615.10
ENST00000619305.1 |
COA6
|
cytochrome c oxidase assembly factor 6 |
| chr15_+_42495617 | 0.14 |
ENST00000564153.5
ENST00000249647.8 ENST00000567094.5 ENST00000566327.5 ENST00000627440.2 ENST00000626061.2 ENST00000568859.5 |
SNAP23
|
synaptosome associated protein 23 |
| chr19_+_7030578 | 0.13 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr6_+_32854179 | 0.13 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr3_-_112975018 | 0.13 |
ENST00000471858.5
ENST00000308611.8 ENST00000295863.4 |
CD200R1
|
CD200 receptor 1 |
| chr7_-_23347704 | 0.13 |
ENST00000619562.4
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr22_-_29766934 | 0.13 |
ENST00000344318.4
|
ZMAT5
|
zinc finger matrin-type 5 |
| chr2_+_27496830 | 0.13 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator |
| chr18_-_500692 | 0.13 |
ENST00000400256.5
|
COLEC12
|
collectin subfamily member 12 |
| chr3_+_44584953 | 0.13 |
ENST00000441021.5
ENST00000322734.2 |
ZNF660
|
zinc finger protein 660 |
| chr22_-_23980469 | 0.13 |
ENST00000404092.5
|
ENSG00000285762.1
|
novel protein, AP000351.4-DDT readtrhough |
| chr11_-_34511710 | 0.13 |
ENST00000620316.4
ENST00000312319.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr8_+_96584920 | 0.13 |
ENST00000521590.5
|
SDC2
|
syndecan 2 |
| chr14_-_52791597 | 0.13 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr7_-_29989774 | 0.13 |
ENST00000242059.10
|
SCRN1
|
secernin 1 |
| chr18_-_5296139 | 0.13 |
ENST00000400143.7
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr1_+_165827786 | 0.13 |
ENST00000642653.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr11_-_111912871 | 0.13 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr12_-_52680398 | 0.12 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr11_+_114296347 | 0.12 |
ENST00000299964.4
|
NNMT
|
nicotinamide N-methyltransferase |
| chr16_+_67173971 | 0.12 |
ENST00000563258.1
ENST00000568146.1 |
NOL3
|
nucleolar protein 3 |
| chr1_-_246193727 | 0.12 |
ENST00000391836.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr2_-_79087986 | 0.12 |
ENST00000305089.8
|
REG1B
|
regenerating family member 1 beta |
| chr17_-_8152380 | 0.12 |
ENST00000317276.9
|
PER1
|
period circadian regulator 1 |
| chr21_-_32813695 | 0.12 |
ENST00000479548.2
ENST00000490358.5 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr19_-_4455292 | 0.12 |
ENST00000394765.7
ENST00000592515.1 |
UBXN6
|
UBX domain protein 6 |
| chr19_+_7049321 | 0.12 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3 like 2 |
| chr5_-_77087245 | 0.12 |
ENST00000255198.3
|
ZBED3
|
zinc finger BED-type containing 3 |
| chr19_-_40716869 | 0.12 |
ENST00000677018.1
ENST00000324464.8 ENST00000594720.6 ENST00000677496.1 |
COQ8B
|
coenzyme Q8B |
| chr22_-_36776067 | 0.12 |
ENST00000417951.6
ENST00000433985.7 ENST00000430701.5 |
IFT27
|
intraflagellar transport 27 |
| chr22_+_36913620 | 0.12 |
ENST00000403662.8
ENST00000262825.9 |
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chr19_-_10334723 | 0.12 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr3_+_9902619 | 0.12 |
ENST00000421412.5
|
IL17RE
|
interleukin 17 receptor E |
| chr8_+_38176802 | 0.12 |
ENST00000287322.5
|
BAG4
|
BAG cochaperone 4 |
| chr22_-_36776147 | 0.12 |
ENST00000340630.9
|
IFT27
|
intraflagellar transport 27 |
| chr19_+_8053000 | 0.12 |
ENST00000390669.7
|
CCL25
|
C-C motif chemokine ligand 25 |
| chrX_-_107716401 | 0.12 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr1_-_27490045 | 0.12 |
ENST00000536657.1
|
WASF2
|
WASP family member 2 |
| chr4_+_37891060 | 0.12 |
ENST00000261439.9
ENST00000508802.5 ENST00000402522.1 |
TBC1D1
|
TBC1 domain family member 1 |
| chr5_+_10441857 | 0.12 |
ENST00000274134.5
|
ROPN1L
|
rhophilin associated tail protein 1 like |
| chr19_-_7040179 | 0.12 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4 |
| chr19_-_58353482 | 0.12 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr2_+_65228122 | 0.12 |
ENST00000542850.2
|
ACTR2
|
actin related protein 2 |
| chr2_+_10368764 | 0.11 |
ENST00000620771.4
|
HPCAL1
|
hippocalcin like 1 |
| chr20_+_36573589 | 0.11 |
ENST00000373872.9
ENST00000650844.1 |
TGIF2
|
TGFB induced factor homeobox 2 |
| chr12_-_47819866 | 0.11 |
ENST00000354334.7
ENST00000430670.5 ENST00000552960.5 ENST00000440293.5 ENST00000080059.12 |
HDAC7
|
histone deacetylase 7 |
| chr9_+_128920966 | 0.11 |
ENST00000428610.5
ENST00000372592.8 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr8_+_124973288 | 0.11 |
ENST00000319286.6
|
ZNF572
|
zinc finger protein 572 |
| chr9_+_122510802 | 0.11 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor family 1 subfamily J member 2 |
| chr3_+_52245721 | 0.11 |
ENST00000323588.9
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent 1M |
| chr3_+_100635598 | 0.11 |
ENST00000475887.1
|
ADGRG7
|
adhesion G protein-coupled receptor G7 |
| chr17_-_59155235 | 0.11 |
ENST00000581068.5
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr19_+_8052752 | 0.11 |
ENST00000315626.6
ENST00000253451.9 |
CCL25
|
C-C motif chemokine ligand 25 |
| chr12_-_7091873 | 0.11 |
ENST00000538050.5
ENST00000536053.6 |
C1R
|
complement C1r |
| chr17_+_7440738 | 0.11 |
ENST00000575398.5
ENST00000575082.5 |
FGF11
|
fibroblast growth factor 11 |
| chrY_-_13986473 | 0.11 |
ENST00000250825.5
|
VCY
|
variable charge Y-linked |
| chr4_+_25312766 | 0.11 |
ENST00000302874.9
ENST00000612982.1 |
ZCCHC4
|
zinc finger CCHC-type containing 4 |
| chr1_-_209806124 | 0.11 |
ENST00000367021.8
ENST00000542854.5 |
IRF6
|
interferon regulatory factor 6 |
| chr6_-_30075767 | 0.11 |
ENST00000244360.8
ENST00000376751.8 |
RNF39
|
ring finger protein 39 |
| chr16_+_56625775 | 0.11 |
ENST00000330439.7
ENST00000568293.1 |
MT1E
|
metallothionein 1E |
| chr4_+_153222402 | 0.11 |
ENST00000676335.1
ENST00000675146.1 |
TRIM2
|
tripartite motif containing 2 |
| chr11_-_124897797 | 0.11 |
ENST00000306534.8
ENST00000533054.5 |
ROBO4
|
roundabout guidance receptor 4 |
| chr1_+_158254414 | 0.11 |
ENST00000289429.6
|
CD1A
|
CD1a molecule |
| chr10_-_27240743 | 0.11 |
ENST00000677901.1
ENST00000677960.1 ENST00000677440.1 ENST00000396271.8 ENST00000677141.1 ENST00000677311.1 ENST00000677667.1 ENST00000677200.1 ENST00000676997.1 ENST00000676511.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_-_86101922 | 0.11 |
ENST00000472236.5
ENST00000641881.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_+_158999963 | 0.11 |
ENST00000566111.5
|
IFI16
|
interferon gamma inducible protein 16 |
| chrX_-_47650488 | 0.11 |
ENST00000247161.7
ENST00000376983.8 ENST00000343894.8 |
ELK1
|
ETS transcription factor ELK1 |
| chr6_+_27247690 | 0.11 |
ENST00000421826.6
ENST00000230582.8 |
PRSS16
|
serine protease 16 |
| chr10_-_27242068 | 0.11 |
ENST00000375901.5
ENST00000412279.1 ENST00000676731.1 ENST00000679220.1 ENST00000678392.1 ENST00000678446.1 ENST00000677441.1 ENST00000375905.8 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr1_+_111619751 | 0.11 |
ENST00000433097.5
ENST00000369709.3 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr3_+_122325237 | 0.11 |
ENST00000264474.4
ENST00000479204.1 |
CSTA
|
cystatin A |
| chr4_+_153222307 | 0.11 |
ENST00000675899.1
ENST00000675611.1 ENST00000674872.1 ENST00000676167.1 |
TRIM2
|
tripartite motif containing 2 |
| chr17_+_8002610 | 0.11 |
ENST00000254854.5
|
GUCY2D
|
guanylate cyclase 2D, retinal |
| chr7_-_75073774 | 0.10 |
ENST00000610322.5
|
RCC1L
|
RCC1 like |
| chr1_-_53220589 | 0.10 |
ENST00000294360.5
|
CZIB
|
CXXC motif containing zinc binding protein |
| chr7_+_118224654 | 0.10 |
ENST00000265224.9
ENST00000486422.1 ENST00000417525.5 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr8_-_132321509 | 0.10 |
ENST00000638588.1
|
KCNQ3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr17_+_31391645 | 0.10 |
ENST00000621161.5
|
RAB11FIP4
|
RAB11 family interacting protein 4 |
| chr2_+_233712905 | 0.10 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr18_-_74457944 | 0.10 |
ENST00000400291.2
|
DIPK1C
|
divergent protein kinase domain 1C |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.1 | 0.4 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.4 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 1.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.3 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.3 | GO:2000360 | positive regulation of female gonad development(GO:2000196) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.6 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.3 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.4 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.1 | 0.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.4 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.6 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.3 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) uracil metabolic process(GO:0019860) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0032712 | regulation of interleukin-3 production(GO:0032672) negative regulation of interleukin-3 production(GO:0032712) negative regulation of granulocyte colony-stimulating factor production(GO:0071656) negative regulation of macrophage colony-stimulating factor production(GO:1901257) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.0 | GO:0019541 | propionate metabolic process(GO:0019541) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0061566 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.0 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.0 | GO:1902232 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:2000124 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.0 | GO:0035498 | carnosine metabolic process(GO:0035498) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0071110 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0060584 | detection of peptidoglycan(GO:0032499) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0044313 | protein K29-linked deubiquitination(GO:0035523) protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.0 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.0 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.0 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.0 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.0 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 0.3 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.3 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0071752 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.5 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.0 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.1 | 0.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.4 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.4 | GO:0089720 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004080 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.0 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |