Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
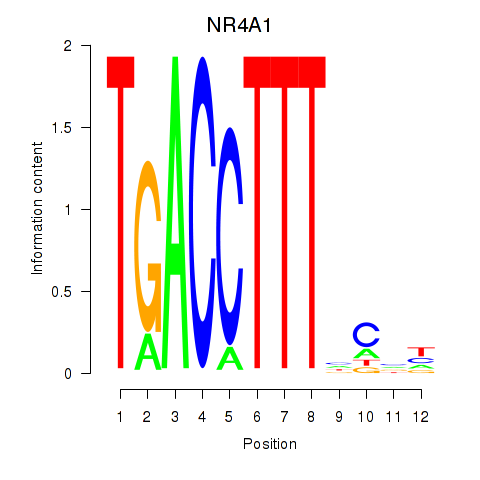
Results for NR4A1
Z-value: 0.28
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.20 | nuclear receptor subfamily 4 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A1 | hg38_v1_chr12_+_52051402_52051459 | -0.10 | 8.2e-01 | Click! |
Activity profile of NR4A1 motif
Sorted Z-values of NR4A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_119673368 | 0.50 |
ENST00000427067.6
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_+_184337591 | 0.22 |
ENST00000383847.7
|
FAM131A
|
family with sequence similarity 131 member A |
| chr20_+_13008919 | 0.21 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr1_-_161132577 | 0.18 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr4_-_158159657 | 0.17 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B |
| chr7_-_38631356 | 0.17 |
ENST00000356264.7
ENST00000325590.9 |
AMPH
|
amphiphysin |
| chr12_+_18242955 | 0.16 |
ENST00000676171.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr20_-_44187153 | 0.15 |
ENST00000372980.4
|
JPH2
|
junctophilin 2 |
| chr3_+_29280945 | 0.14 |
ENST00000434693.6
|
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr6_-_152168291 | 0.14 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr6_-_152168349 | 0.14 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr6_+_69232406 | 0.13 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr10_-_59906509 | 0.13 |
ENST00000263102.7
|
CCDC6
|
coiled-coil domain containing 6 |
| chr15_+_75043263 | 0.12 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr17_-_7404039 | 0.12 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr4_+_70242583 | 0.12 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr2_+_161160299 | 0.12 |
ENST00000440506.5
ENST00000429217.5 ENST00000406287.5 ENST00000402568.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr11_-_111923722 | 0.12 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr19_+_6372757 | 0.12 |
ENST00000245812.8
|
ALKBH7
|
alkB homolog 7 |
| chr2_-_177392673 | 0.12 |
ENST00000447413.1
ENST00000397057.6 ENST00000456746.5 ENST00000464747.5 |
ENSG00000213963.6
NFE2L2
|
novel transcript nuclear factor, erythroid 2 like 2 |
| chr1_-_161132659 | 0.12 |
ENST00000368006.8
ENST00000490843.6 ENST00000545495.5 |
DEDD
|
death effector domain containing |
| chr12_+_32502114 | 0.11 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr12_+_21526287 | 0.11 |
ENST00000256969.7
|
SPX
|
spexin hormone |
| chr7_-_105876575 | 0.11 |
ENST00000318724.8
ENST00000419735.8 |
ATXN7L1
|
ataxin 7 like 1 |
| chr6_-_152637351 | 0.10 |
ENST00000367255.10
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr20_+_38962299 | 0.10 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr4_-_88823165 | 0.10 |
ENST00000508369.5
|
FAM13A
|
family with sequence similarity 13 member A |
| chr20_+_17699942 | 0.09 |
ENST00000427254.1
ENST00000377805.7 |
BANF2
|
BANF family member 2 |
| chr19_+_17305801 | 0.09 |
ENST00000602206.1
ENST00000252602.2 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr12_+_95858928 | 0.09 |
ENST00000266735.9
ENST00000553192.5 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr20_-_44187093 | 0.09 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chr1_+_61404076 | 0.09 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr4_-_88823214 | 0.09 |
ENST00000513837.5
ENST00000503556.5 |
FAM13A
|
family with sequence similarity 13 member A |
| chr6_+_44158807 | 0.08 |
ENST00000532171.5
ENST00000398776.2 |
CAPN11
|
calpain 11 |
| chr2_+_161160420 | 0.08 |
ENST00000392749.7
ENST00000405852.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr4_+_99574812 | 0.08 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr20_+_11917859 | 0.08 |
ENST00000618296.4
ENST00000378226.7 |
BTBD3
|
BTB domain containing 3 |
| chr10_+_35195843 | 0.08 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr2_+_233307806 | 0.08 |
ENST00000447536.5
ENST00000409110.6 |
SAG
|
S-antigen visual arrestin |
| chr13_-_30306997 | 0.08 |
ENST00000380617.7
ENST00000441394.1 |
KATNAL1
|
katanin catalytic subunit A1 like 1 |
| chr1_+_192809031 | 0.08 |
ENST00000235382.7
|
RGS2
|
regulator of G protein signaling 2 |
| chr11_+_116829898 | 0.07 |
ENST00000227667.8
ENST00000375345.3 |
APOC3
|
apolipoprotein C3 |
| chr2_+_177392734 | 0.07 |
ENST00000680770.1
ENST00000637633.2 ENST00000679459.1 ENST00000409888.1 ENST00000264167.11 ENST00000642466.2 |
AGPS
|
alkylglycerone phosphate synthase |
| chr8_+_98426943 | 0.07 |
ENST00000287042.5
|
KCNS2
|
potassium voltage-gated channel modifier subfamily S member 2 |
| chr17_-_7219813 | 0.07 |
ENST00000399510.8
ENST00000648172.8 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr14_-_77028663 | 0.07 |
ENST00000238647.5
|
IRF2BPL
|
interferon regulatory factor 2 binding protein like |
| chr1_+_160205374 | 0.07 |
ENST00000368077.5
ENST00000360472.9 |
PEA15
|
proliferation and apoptosis adaptor protein 15 |
| chr1_+_160205411 | 0.06 |
ENST00000368076.1
|
PEA15
|
proliferation and apoptosis adaptor protein 15 |
| chr3_-_115147237 | 0.06 |
ENST00000357258.8
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_+_54415427 | 0.06 |
ENST00000301194.8
ENST00000376530.8 ENST00000445095.5 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr3_-_48898813 | 0.06 |
ENST00000319017.5
ENST00000430379.5 |
SLC25A20
|
solute carrier family 25 member 20 |
| chr3_-_115147277 | 0.06 |
ENST00000675478.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr8_-_91040814 | 0.06 |
ENST00000520014.1
ENST00000285419.8 |
PIP4P2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr20_-_45857196 | 0.05 |
ENST00000457981.5
ENST00000426915.1 ENST00000217455.9 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr12_+_109139397 | 0.05 |
ENST00000377854.9
ENST00000377848.7 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr1_+_165631199 | 0.05 |
ENST00000367888.8
ENST00000367889.8 ENST00000367885.5 ENST00000367884.6 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr11_-_113706292 | 0.05 |
ENST00000544634.5
ENST00000539732.5 ENST00000538770.1 ENST00000536856.5 ENST00000299882.11 ENST00000544476.1 |
TMPRSS5
|
transmembrane serine protease 5 |
| chr6_-_43627302 | 0.05 |
ENST00000307114.11
|
GTPBP2
|
GTP binding protein 2 |
| chr17_-_2266131 | 0.05 |
ENST00000570606.5
ENST00000354901.8 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr12_-_76878985 | 0.05 |
ENST00000547435.1
ENST00000552330.5 ENST00000311083.10 ENST00000546966.5 |
CSRP2
|
cysteine and glycine rich protein 2 |
| chr2_+_176157293 | 0.05 |
ENST00000683222.1
|
HOXD3
|
homeobox D3 |
| chr4_-_88823306 | 0.05 |
ENST00000395002.6
|
FAM13A
|
family with sequence similarity 13 member A |
| chr7_-_6009072 | 0.05 |
ENST00000642456.1
ENST00000642292.1 ENST00000441476.6 |
PMS2
|
PMS1 homolog 2, mismatch repair system component |
| chr17_+_7219857 | 0.05 |
ENST00000583312.5
ENST00000350303.9 ENST00000584103.5 ENST00000356839.10 ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase very long chain |
| chr11_-_113706330 | 0.05 |
ENST00000540540.5
ENST00000538955.5 ENST00000545579.6 |
TMPRSS5
|
transmembrane serine protease 5 |
| chr12_+_6535278 | 0.05 |
ENST00000396858.5
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr4_-_56387422 | 0.05 |
ENST00000513376.5
ENST00000205214.11 ENST00000502617.1 ENST00000451613.5 ENST00000602986.5 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr7_-_105876477 | 0.05 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7 like 1 |
| chr16_-_31150058 | 0.05 |
ENST00000569305.1
ENST00000268281.9 ENST00000418068.6 |
PRSS36
|
serine protease 36 |
| chr19_-_40750302 | 0.05 |
ENST00000598485.6
ENST00000378313.7 ENST00000470681.5 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr10_-_73874461 | 0.05 |
ENST00000305762.11
|
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma |
| chr12_+_57941499 | 0.05 |
ENST00000300145.4
|
ATP23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr3_-_185821092 | 0.04 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr2_-_25252072 | 0.04 |
ENST00000683760.1
|
DNMT3A
|
DNA methyltransferase 3 alpha |
| chr12_+_53985138 | 0.04 |
ENST00000303460.5
|
HOXC10
|
homeobox C10 |
| chr7_-_73522278 | 0.04 |
ENST00000404251.1
ENST00000339594.9 |
BAZ1B
|
bromodomain adjacent to zinc finger domain 1B |
| chr20_+_17699960 | 0.04 |
ENST00000246090.6
|
BANF2
|
BANF family member 2 |
| chr17_+_4951080 | 0.04 |
ENST00000521811.5
ENST00000323997.10 ENST00000522249.5 ENST00000519584.5 ENST00000519602.6 |
ENO3
|
enolase 3 |
| chrX_-_129523436 | 0.04 |
ENST00000371123.5
ENST00000371121.5 ENST00000371122.8 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr1_+_40247926 | 0.04 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr5_-_127073467 | 0.04 |
ENST00000607731.5
ENST00000509733.7 ENST00000296662.10 ENST00000535381.6 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr20_+_58891981 | 0.03 |
ENST00000488652.6
ENST00000476935.6 ENST00000492907.6 ENST00000603546.2 |
GNAS
|
GNAS complex locus |
| chr3_-_113746059 | 0.03 |
ENST00000477813.5
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr2_-_24972032 | 0.03 |
ENST00000534855.5
|
DNAJC27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr6_+_31927683 | 0.03 |
ENST00000456570.5
|
ENSG00000244255.5
|
novel complement component 2 (C2) and complement factor B (CFB) protein |
| chr3_+_138621225 | 0.03 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_+_63636481 | 0.03 |
ENST00000509330.5
|
PHF3
|
PHD finger protein 3 |
| chr16_+_53207981 | 0.03 |
ENST00000565803.2
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr14_-_103921494 | 0.03 |
ENST00000557040.5
ENST00000414262.6 ENST00000555030.5 ENST00000554713.2 ENST00000286953.8 ENST00000553430.5 |
ATP5MJ
|
ATP synthase membrane subunit j |
| chr3_-_157503574 | 0.03 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr12_+_21372899 | 0.03 |
ENST00000240652.8
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr1_+_15736251 | 0.03 |
ENST00000294454.6
|
SLC25A34
|
solute carrier family 25 member 34 |
| chr14_+_35122537 | 0.03 |
ENST00000604948.5
ENST00000321130.14 ENST00000534898.9 ENST00000605201.1 ENST00000250377.11 |
PRORP
|
protein only RNase P catalytic subunit |
| chrX_+_2828808 | 0.03 |
ENST00000381163.7
|
GYG2
|
glycogenin 2 |
| chr13_-_32538683 | 0.03 |
ENST00000674456.1
ENST00000504114.5 |
N4BP2L2
|
NEDD4 binding protein 2 like 2 |
| chr7_-_6009019 | 0.03 |
ENST00000382321.5
ENST00000265849.12 |
PMS2
|
PMS1 homolog 2, mismatch repair system component |
| chr18_+_46174055 | 0.03 |
ENST00000615553.1
|
C18orf25
|
chromosome 18 open reading frame 25 |
| chr4_-_40630588 | 0.03 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_+_4951758 | 0.02 |
ENST00000518175.1
|
ENO3
|
enolase 3 |
| chr3_-_157503375 | 0.02 |
ENST00000362010.7
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr3_+_138621207 | 0.02 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr7_-_81770039 | 0.02 |
ENST00000222390.11
ENST00000453411.6 ENST00000457544.7 ENST00000444829.7 |
HGF
|
hepatocyte growth factor |
| chr18_-_63319729 | 0.02 |
ENST00000333681.5
|
BCL2
|
BCL2 apoptosis regulator |
| chr3_-_157503339 | 0.02 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr12_-_16605939 | 0.02 |
ENST00000541295.5
ENST00000535535.5 |
LMO3
|
LIM domain only 3 |
| chr3_-_44477639 | 0.02 |
ENST00000396077.8
|
ZNF445
|
zinc finger protein 445 |
| chr14_+_21918161 | 0.02 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr2_-_24971900 | 0.02 |
ENST00000264711.7
|
DNAJC27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr22_+_24495242 | 0.02 |
ENST00000382760.2
ENST00000326010.10 |
UPB1
|
beta-ureidopropionase 1 |
| chr10_+_99782628 | 0.02 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chrX_+_2828921 | 0.02 |
ENST00000398806.8
|
GYG2
|
glycogenin 2 |
| chr12_+_93569814 | 0.02 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr18_-_27143024 | 0.02 |
ENST00000581714.5
|
CHST9
|
carbohydrate sulfotransferase 9 |
| chr15_+_36594868 | 0.01 |
ENST00000566807.5
ENST00000643612.1 ENST00000567389.5 ENST00000562877.5 |
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr1_+_69567906 | 0.01 |
ENST00000651989.2
|
LRRC7
|
leucine rich repeat containing 7 |
| chr19_+_14028148 | 0.01 |
ENST00000431365.3
ENST00000585987.1 |
RLN3
|
relaxin 3 |
| chrX_-_132961390 | 0.01 |
ENST00000370836.6
ENST00000521489.5 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr1_+_50103903 | 0.01 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr3_-_139044892 | 0.01 |
ENST00000413199.1
|
PRR23C
|
proline rich 23C |
| chr17_+_42998779 | 0.01 |
ENST00000586277.5
|
RPL27
|
ribosomal protein L27 |
| chr17_+_42998379 | 0.01 |
ENST00000253788.12
ENST00000589913.6 |
RPL27
|
ribosomal protein L27 |
| chr6_+_30626842 | 0.01 |
ENST00000318999.11
ENST00000376485.8 ENST00000330083.6 ENST00000319027.9 ENST00000376483.8 ENST00000329992.12 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr12_-_16606102 | 0.01 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3 |
| chr6_+_31927703 | 0.01 |
ENST00000418949.6
ENST00000299367.10 ENST00000383177.7 ENST00000477310.1 |
C2
ENSG00000244255.5
|
complement C2 novel complement component 2 (C2) and complement factor B (CFB) protein |
| chr17_+_42998264 | 0.01 |
ENST00000589037.5
|
RPL27
|
ribosomal protein L27 |
| chr14_-_35121950 | 0.01 |
ENST00000554361.5
ENST00000261475.10 |
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr17_-_31297231 | 0.01 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr3_+_101724602 | 0.01 |
ENST00000341893.8
|
CEP97
|
centrosomal protein 97 |
| chr7_+_121328991 | 0.01 |
ENST00000222462.3
|
WNT16
|
Wnt family member 16 |
| chr2_-_206159410 | 0.01 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr3_+_113747022 | 0.01 |
ENST00000273398.8
ENST00000496747.5 ENST00000475322.1 |
ATP6V1A
|
ATPase H+ transporting V1 subunit A |
| chr10_-_72954790 | 0.01 |
ENST00000373032.4
|
PLA2G12B
|
phospholipase A2 group XIIB |
| chr19_-_11529116 | 0.01 |
ENST00000592312.5
ENST00000590480.1 ENST00000585318.5 ENST00000270517.12 ENST00000252440.11 ENST00000417981.6 |
ECSIT
|
ECSIT signaling integrator |
| chr2_+_206159884 | 0.01 |
ENST00000392222.7
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_13749408 | 0.01 |
ENST00000343137.8
ENST00000503842.5 ENST00000407521.7 ENST00000505823.5 |
PRDM2
|
PR/SET domain 2 |
| chr12_-_16606795 | 0.00 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3 |
| chr4_-_73223082 | 0.00 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr7_-_102616692 | 0.00 |
ENST00000521076.5
ENST00000462172.5 ENST00000522801.5 ENST00000262940.12 ENST00000449970.6 |
RASA4
|
RAS p21 protein activator 4 |
| chr6_-_33803980 | 0.00 |
ENST00000507738.1
ENST00000266003.9 ENST00000430124.7 |
MLN
|
motilin |
| chr1_+_212565334 | 0.00 |
ENST00000366981.8
ENST00000366987.6 |
ATF3
|
activating transcription factor 3 |
| chr20_+_3071618 | 0.00 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chr7_-_107803215 | 0.00 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr1_+_228208024 | 0.00 |
ENST00000570156.7
ENST00000680850.1 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr3_-_30894661 | 0.00 |
ENST00000282538.10
ENST00000454381.3 |
GADL1
|
glutamate decarboxylase like 1 |
| chr11_+_66857056 | 0.00 |
ENST00000309602.5
ENST00000393952.3 |
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr16_+_55655960 | 0.00 |
ENST00000568943.6
|
SLC6A2
|
solute carrier family 6 member 2 |
| chr16_-_30029429 | 0.00 |
ENST00000564806.1
|
TLCD3B
|
TLC domain containing 3B |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0060621 | regulation of high-density lipoprotein particle clearance(GO:0010982) negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.0 | 0.0 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |