Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
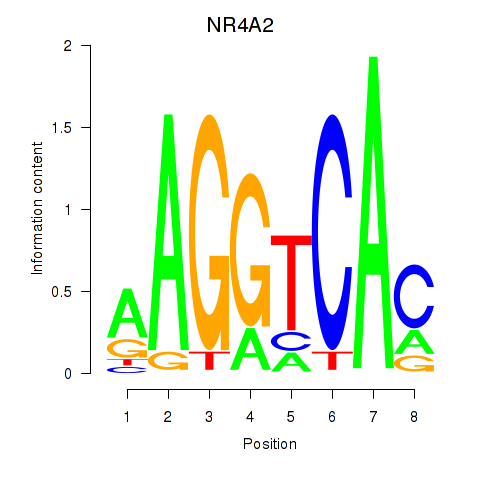
Results for NR4A2
Z-value: 0.74
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.15 | nuclear receptor subfamily 4 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A2 | hg38_v1_chr2_-_156332694_156332728 | -0.93 | 8.8e-04 | Click! |
Activity profile of NR4A2 motif
Sorted Z-values of NR4A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43593054 | 0.77 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr15_+_43692886 | 0.66 |
ENST00000434505.5
ENST00000411750.5 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr20_+_59604527 | 0.66 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr16_-_68236069 | 0.64 |
ENST00000473183.7
ENST00000565858.5 |
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr5_+_66828762 | 0.56 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_2711633 | 0.41 |
ENST00000435359.5
|
CLUH
|
clustered mitochondria homolog |
| chr3_-_151316795 | 0.40 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr2_-_110115811 | 0.36 |
ENST00000272462.3
|
MALL
|
mal, T cell differentiation protein like |
| chr4_+_20253500 | 0.33 |
ENST00000622093.4
ENST00000273739.9 |
SLIT2
|
slit guidance ligand 2 |
| chr10_-_75235917 | 0.32 |
ENST00000469299.1
ENST00000372538.8 |
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr17_-_2711736 | 0.32 |
ENST00000651024.2
ENST00000576885.5 ENST00000574426.7 |
CLUH
|
clustered mitochondria homolog |
| chr10_+_79347491 | 0.31 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr2_+_219627394 | 0.31 |
ENST00000373760.6
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr4_-_102345469 | 0.30 |
ENST00000356736.5
ENST00000682932.1 |
SLC39A8
|
solute carrier family 39 member 8 |
| chr11_-_66371728 | 0.29 |
ENST00000357440.7
ENST00000619145.4 |
SLC29A2
|
solute carrier family 29 member 2 |
| chr17_-_29176752 | 0.28 |
ENST00000533112.5
|
MYO18A
|
myosin XVIIIA |
| chr7_-_81770122 | 0.27 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr10_+_79347460 | 0.25 |
ENST00000225174.8
|
PPIF
|
peptidylprolyl isomerase F |
| chr16_-_58295019 | 0.24 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr16_-_58294976 | 0.24 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chr1_-_115768702 | 0.23 |
ENST00000261448.6
|
CASQ2
|
calsequestrin 2 |
| chr20_+_1895365 | 0.23 |
ENST00000358771.5
|
SIRPA
|
signal regulatory protein alpha |
| chr4_-_40515967 | 0.23 |
ENST00000381795.10
|
RBM47
|
RNA binding motif protein 47 |
| chr1_+_9588860 | 0.22 |
ENST00000340381.11
ENST00000340305.9 |
TMEM201
|
transmembrane protein 201 |
| chr16_+_85908988 | 0.22 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr11_+_76860859 | 0.21 |
ENST00000679754.1
ENST00000534206.5 ENST00000680583.1 ENST00000532485.6 ENST00000526597.5 ENST00000533873.1 |
ACER3
|
alkaline ceramidase 3 |
| chr4_-_138242325 | 0.21 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr21_+_41167774 | 0.20 |
ENST00000328735.10
ENST00000347667.5 |
BACE2
|
beta-secretase 2 |
| chr8_+_144095054 | 0.19 |
ENST00000318911.5
|
CYC1
|
cytochrome c1 |
| chr4_-_1856090 | 0.19 |
ENST00000302787.3
|
LETM1
|
leucine zipper and EF-hand containing transmembrane protein 1 |
| chr7_+_73830988 | 0.19 |
ENST00000340958.4
|
CLDN4
|
claudin 4 |
| chr8_-_130016414 | 0.19 |
ENST00000401979.6
ENST00000517654.5 ENST00000522361.1 ENST00000518167.5 |
CYRIB
|
CYFIP related Rac1 interactor B |
| chr7_+_48088596 | 0.18 |
ENST00000416681.5
ENST00000331803.8 ENST00000432131.5 |
UPP1
|
uridine phosphorylase 1 |
| chr7_+_48089257 | 0.18 |
ENST00000436673.5
ENST00000395564.9 |
UPP1
|
uridine phosphorylase 1 |
| chr21_+_41168142 | 0.18 |
ENST00000330333.11
|
BACE2
|
beta-secretase 2 |
| chr8_-_130016395 | 0.18 |
ENST00000523509.5
|
CYRIB
|
CYFIP related Rac1 interactor B |
| chr14_-_21023954 | 0.18 |
ENST00000554094.5
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21024092 | 0.17 |
ENST00000554398.5
|
NDRG2
|
NDRG family member 2 |
| chr12_+_132490140 | 0.17 |
ENST00000680143.1
|
FBRSL1
|
fibrosin like 1 |
| chr11_-_9314564 | 0.17 |
ENST00000611268.4
ENST00000528080.6 ENST00000527813.5 ENST00000533723.1 |
TMEM41B
|
transmembrane protein 41B |
| chr9_-_39239174 | 0.17 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr1_+_236686454 | 0.17 |
ENST00000542672.6
ENST00000366578.6 ENST00000682015.1 ENST00000651275.1 |
ACTN2
|
actinin alpha 2 |
| chr13_-_52011337 | 0.17 |
ENST00000400366.6
ENST00000400370.8 ENST00000634844.1 ENST00000673772.1 ENST00000418097.7 ENST00000242839.10 ENST00000344297.9 ENST00000448424.7 |
ATP7B
|
ATPase copper transporting beta |
| chr19_+_44891206 | 0.17 |
ENST00000405636.6
ENST00000252487.9 ENST00000592434.5 ENST00000589649.1 ENST00000426677.7 |
TOMM40
|
translocase of outer mitochondrial membrane 40 |
| chr7_+_112450451 | 0.16 |
ENST00000429071.5
ENST00000403825.8 ENST00000675268.1 |
IFRD1
ENSG00000288634.1
|
interferon related developmental regulator 1 novel protein |
| chr7_-_73522278 | 0.15 |
ENST00000404251.1
ENST00000339594.9 |
BAZ1B
|
bromodomain adjacent to zinc finger domain 1B |
| chr8_-_130016622 | 0.15 |
ENST00000518283.5
ENST00000519110.5 |
CYRIB
|
CYFIP related Rac1 interactor B |
| chr8_+_22053543 | 0.14 |
ENST00000519850.5
ENST00000381470.7 |
DMTN
|
dematin actin binding protein |
| chr8_+_144358974 | 0.14 |
ENST00000526891.2
|
SLC52A2
|
solute carrier family 52 member 2 |
| chr18_-_12377200 | 0.14 |
ENST00000269143.8
|
AFG3L2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chr9_-_114682172 | 0.14 |
ENST00000436752.3
|
TEX48
|
testis expressed 48 |
| chr11_+_45805108 | 0.14 |
ENST00000530471.1
ENST00000314134.4 |
SLC35C1
|
solute carrier family 35 member C1 |
| chr9_-_114682061 | 0.13 |
ENST00000612244.5
|
TEX48
|
testis expressed 48 |
| chr10_-_100081854 | 0.13 |
ENST00000370418.8
|
CPN1
|
carboxypeptidase N subunit 1 |
| chr1_+_174447944 | 0.13 |
ENST00000367685.5
|
GPR52
|
G protein-coupled receptor 52 |
| chr14_+_21030201 | 0.13 |
ENST00000321760.11
ENST00000460647.6 ENST00000530140.6 ENST00000472458.5 |
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chrX_+_154411519 | 0.13 |
ENST00000612460.5
ENST00000601016.6 ENST00000613002.4 ENST00000475699.6 ENST00000612012.5 ENST00000369776.8 ENST00000439735.2 ENST00000616020.5 ENST00000652358.1 ENST00000652390.1 |
TAZ
|
tafazzin |
| chr7_-_126533850 | 0.13 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr8_-_143944737 | 0.13 |
ENST00000398774.6
|
PLEC
|
plectin |
| chr15_+_59438149 | 0.12 |
ENST00000288228.10
ENST00000559628.5 ENST00000557914.5 ENST00000560474.5 |
FAM81A
|
family with sequence similarity 81 member A |
| chr7_+_39623886 | 0.12 |
ENST00000436179.1
|
RALA
|
RAS like proto-oncogene A |
| chr8_-_18083184 | 0.12 |
ENST00000636269.1
|
ASAH1
|
N-acylsphingosine amidohydrolase 1 |
| chr1_-_157138474 | 0.12 |
ENST00000326786.4
|
ETV3
|
ETS variant transcription factor 3 |
| chr4_-_151015263 | 0.12 |
ENST00000510413.5
ENST00000507224.5 ENST00000651943.2 |
LRBA
|
LPS responsive beige-like anchor protein |
| chr11_+_73950985 | 0.12 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr7_+_23598144 | 0.11 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr7_+_37920602 | 0.11 |
ENST00000199448.9
ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr6_-_106629472 | 0.11 |
ENST00000369063.8
|
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr19_+_1407731 | 0.11 |
ENST00000592453.2
|
DAZAP1
|
DAZ associated protein 1 |
| chr17_-_676348 | 0.11 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr1_+_6034677 | 0.11 |
ENST00000668559.1
|
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr4_-_151015713 | 0.11 |
ENST00000357115.9
|
LRBA
|
LPS responsive beige-like anchor protein |
| chr4_-_110198650 | 0.11 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_+_80197493 | 0.11 |
ENST00000415738.3
|
PRDM8
|
PR/SET domain 8 |
| chr22_-_37484505 | 0.11 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_110198579 | 0.10 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr22_-_29388530 | 0.10 |
ENST00000357586.7
ENST00000432560.6 ENST00000405198.6 ENST00000317368.11 |
AP1B1
|
adaptor related protein complex 1 subunit beta 1 |
| chr1_+_6034980 | 0.10 |
ENST00000378092.6
ENST00000472700.7 |
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr2_+_119431846 | 0.10 |
ENST00000306406.5
|
TMEM37
|
transmembrane protein 37 |
| chr17_+_7630094 | 0.10 |
ENST00000441599.6
ENST00000380450.9 ENST00000416273.7 ENST00000575903.5 ENST00000571153.5 ENST00000575618.5 ENST00000576152.1 ENST00000576830.5 |
SHBG
|
sex hormone binding globulin |
| chr10_-_49762335 | 0.09 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr1_-_225941383 | 0.09 |
ENST00000420304.6
|
LEFTY2
|
left-right determination factor 2 |
| chr5_+_218241 | 0.09 |
ENST00000617470.4
ENST00000504309.5 ENST00000510361.5 ENST00000264932.11 |
SDHA
|
succinate dehydrogenase complex flavoprotein subunit A |
| chr19_-_15125751 | 0.09 |
ENST00000263383.8
|
ILVBL
|
ilvB acetolactate synthase like |
| chr1_+_212565334 | 0.09 |
ENST00000366981.8
ENST00000366987.6 |
ATF3
|
activating transcription factor 3 |
| chr16_+_56730099 | 0.09 |
ENST00000563858.5
ENST00000566315.5 ENST00000308159.10 ENST00000569842.5 ENST00000569863.5 |
NUP93
|
nucleoporin 93 |
| chr6_+_106629594 | 0.09 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA amidotransferase subunit QRSL1 |
| chr4_-_98929092 | 0.09 |
ENST00000280892.10
ENST00000511644.5 ENST00000504432.5 ENST00000450253.7 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr13_-_113864062 | 0.08 |
ENST00000327773.7
|
GAS6
|
growth arrest specific 6 |
| chr1_+_43337803 | 0.08 |
ENST00000372470.9
ENST00000413998.7 |
MPL
|
MPL proto-oncogene, thrombopoietin receptor |
| chr4_+_24795560 | 0.08 |
ENST00000382120.4
|
SOD3
|
superoxide dismutase 3 |
| chr3_-_42701513 | 0.08 |
ENST00000310417.9
|
HHATL
|
hedgehog acyltransferase like |
| chr12_+_10010627 | 0.08 |
ENST00000338896.11
ENST00000396502.5 |
CLEC12B
|
C-type lectin domain family 12 member B |
| chr15_+_64911869 | 0.08 |
ENST00000319580.13
ENST00000496660.5 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr20_+_19758245 | 0.08 |
ENST00000255006.12
|
RIN2
|
Ras and Rab interactor 2 |
| chr4_+_41612892 | 0.08 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_197298558 | 0.08 |
ENST00000656944.1
ENST00000346964.6 ENST00000448528.6 ENST00000655488.1 ENST00000357674.9 ENST00000667157.1 ENST00000661336.1 ENST00000654737.1 ENST00000659716.1 ENST00000657381.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr13_-_103066411 | 0.08 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2 |
| chr19_-_55063170 | 0.08 |
ENST00000610356.4
|
RDH13
|
retinol dehydrogenase 13 |
| chr10_-_49942027 | 0.08 |
ENST00000616448.2
|
PARG
|
poly(ADP-ribose) glycohydrolase |
| chr3_-_113746218 | 0.08 |
ENST00000497255.1
ENST00000240922.8 ENST00000478020.1 ENST00000493900.5 |
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr7_+_39623547 | 0.08 |
ENST00000005257.7
|
RALA
|
RAS like proto-oncogene A |
| chr3_+_9703826 | 0.08 |
ENST00000613455.4
ENST00000383831.7 ENST00000383832.8 |
CPNE9
|
copine family member 9 |
| chr12_+_120438107 | 0.08 |
ENST00000229379.3
ENST00000551806.1 |
COX6A1
ENSG00000111780.8
|
cytochrome c oxidase subunit 6A1 novel protein |
| chr2_+_102761963 | 0.08 |
ENST00000640575.2
ENST00000412401.3 |
TMEM182
|
transmembrane protein 182 |
| chr1_-_225941212 | 0.08 |
ENST00000366820.10
|
LEFTY2
|
left-right determination factor 2 |
| chr16_-_81198766 | 0.07 |
ENST00000526632.5
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chr5_-_132011811 | 0.07 |
ENST00000379255.5
ENST00000430403.5 ENST00000357096.5 |
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
| chr6_-_39322540 | 0.07 |
ENST00000425054.6
ENST00000373227.8 ENST00000373229.9 |
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr3_+_57556244 | 0.07 |
ENST00000311180.9
ENST00000487257.1 |
PDE12
|
phosphodiesterase 12 |
| chr19_-_56121223 | 0.07 |
ENST00000587279.1
ENST00000610935.2 |
ZNF787
|
zinc finger protein 787 |
| chr19_-_7943648 | 0.07 |
ENST00000597926.1
ENST00000270538.8 |
TIMM44
|
translocase of inner mitochondrial membrane 44 |
| chr3_+_52779916 | 0.07 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr8_+_135457442 | 0.07 |
ENST00000355849.10
|
KHDRBS3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr6_-_31660735 | 0.07 |
ENST00000375911.2
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr17_-_58280928 | 0.07 |
ENST00000225275.4
|
MPO
|
myeloperoxidase |
| chr19_-_35563653 | 0.07 |
ENST00000262623.4
|
ATP4A
|
ATPase H+/K+ transporting subunit alpha |
| chr19_+_589873 | 0.07 |
ENST00000251287.3
|
HCN2
|
hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2 |
| chr1_-_205935822 | 0.07 |
ENST00000340781.8
|
SLC26A9
|
solute carrier family 26 member 9 |
| chr11_+_3855629 | 0.07 |
ENST00000526596.2
ENST00000300737.8 ENST00000616714.4 |
STIM1
|
stromal interaction molecule 1 |
| chr15_-_40828699 | 0.07 |
ENST00000299174.10
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1 regulatory inhibitor subunit 14D |
| chr2_+_233718734 | 0.06 |
ENST00000373409.8
|
UGT1A4
|
UDP glucuronosyltransferase family 1 member A4 |
| chr3_-_167734465 | 0.06 |
ENST00000487947.6
|
PDCD10
|
programmed cell death 10 |
| chr19_+_1407653 | 0.06 |
ENST00000587079.5
|
DAZAP1
|
DAZ associated protein 1 |
| chr2_-_197499826 | 0.06 |
ENST00000439605.2
ENST00000388968.8 ENST00000418022.2 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr1_+_155209213 | 0.06 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr1_+_44674688 | 0.06 |
ENST00000418644.5
ENST00000458657.6 ENST00000535358.6 ENST00000441519.5 ENST00000445071.5 |
ARMH1
|
armadillo like helical domain containing 1 |
| chr2_-_206159509 | 0.06 |
ENST00000423725.5
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr20_+_54475584 | 0.06 |
ENST00000262593.10
|
DOK5
|
docking protein 5 |
| chrX_+_153517626 | 0.06 |
ENST00000263519.5
|
ATP2B3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr7_+_112423137 | 0.06 |
ENST00000005558.8
ENST00000621379.4 |
IFRD1
|
interferon related developmental regulator 1 |
| chr2_+_233712905 | 0.06 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chrX_+_44873552 | 0.06 |
ENST00000683021.1
|
KDM6A
|
lysine demethylase 6A |
| chr3_-_167734510 | 0.06 |
ENST00000475915.6
ENST00000462725.6 ENST00000461494.5 |
PDCD10
|
programmed cell death 10 |
| chr1_-_20661356 | 0.06 |
ENST00000602624.7
ENST00000464364.1 |
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase non-catalytic subunit |
| chrX_+_44873169 | 0.06 |
ENST00000675577.1
ENST00000674867.1 ENST00000674586.1 ENST00000382899.9 ENST00000536777.6 ENST00000543216.6 ENST00000377967.9 ENST00000611820.5 |
KDM6A
|
lysine demethylase 6A |
| chr3_-_42702778 | 0.06 |
ENST00000457462.5
ENST00000441594.6 |
HHATL
|
hedgehog acyltransferase like |
| chr22_+_31122923 | 0.06 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr2_+_219442023 | 0.06 |
ENST00000431523.5
ENST00000396698.5 |
SPEG
|
striated muscle enriched protein kinase |
| chr1_-_157138388 | 0.06 |
ENST00000368192.9
|
ETV3
|
ETS variant transcription factor 3 |
| chr9_+_110668779 | 0.05 |
ENST00000416899.7
ENST00000374448.9 |
MUSK
|
muscle associated receptor tyrosine kinase |
| chr9_-_133479075 | 0.05 |
ENST00000414172.1
ENST00000371897.8 ENST00000371899.9 |
SLC2A6
|
solute carrier family 2 member 6 |
| chrX_+_70452286 | 0.05 |
ENST00000374355.7
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr11_+_66857056 | 0.05 |
ENST00000309602.5
ENST00000393952.3 |
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_-_49049111 | 0.05 |
ENST00000448456.4
|
CGB8
|
chorionic gonadotropin subunit beta 8 |
| chr3_-_172711166 | 0.05 |
ENST00000538775.5
ENST00000543711.5 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr20_+_33168148 | 0.05 |
ENST00000354932.6
|
BPIFA2
|
BPI fold containing family A member 2 |
| chr10_-_14838003 | 0.05 |
ENST00000465530.2
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr2_-_197499857 | 0.05 |
ENST00000428204.6
ENST00000678170.1 ENST00000676933.1 ENST00000678621.1 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr12_-_69699331 | 0.05 |
ENST00000548658.1
ENST00000476098.5 |
BEST3
|
bestrophin 3 |
| chr5_+_96936071 | 0.05 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase |
| chr12_+_93569814 | 0.05 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr17_+_81703356 | 0.05 |
ENST00000333676.8
ENST00000571730.1 |
MRPL12
ENSG00000262660.1
|
mitochondrial ribosomal protein L12 novel protein |
| chr5_-_150412743 | 0.05 |
ENST00000353334.11
ENST00000009530.12 ENST00000377795.7 |
CD74
|
CD74 molecule |
| chr12_-_108561157 | 0.05 |
ENST00000228284.8
ENST00000431469.6 ENST00000546815.6 |
SART3
|
spliceosome associated factor 3, U4/U6 recycling protein |
| chr19_+_1407517 | 0.05 |
ENST00000336761.10
ENST00000233078.9 ENST00000592522.5 |
DAZAP1
|
DAZ associated protein 1 |
| chr10_-_126670686 | 0.05 |
ENST00000488181.3
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr16_-_1884231 | 0.05 |
ENST00000563416.3
ENST00000633813.1 ENST00000470044.5 |
LINC00254
MEIOB
|
long intergenic non-protein coding RNA 254 meiosis specific with OB-fold |
| chr7_-_81770039 | 0.05 |
ENST00000222390.11
ENST00000453411.6 ENST00000457544.7 ENST00000444829.7 |
HGF
|
hepatocyte growth factor |
| chr10_-_48914232 | 0.05 |
ENST00000374160.7
|
LRRC18
|
leucine rich repeat containing 18 |
| chr9_-_135961310 | 0.05 |
ENST00000371756.4
|
UBAC1
|
UBA domain containing 1 |
| chr12_-_69699455 | 0.05 |
ENST00000266661.8
|
BEST3
|
bestrophin 3 |
| chrY_-_13479938 | 0.05 |
ENST00000382893.2
ENST00000382896.9 ENST00000545955.6 |
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr8_+_133191060 | 0.04 |
ENST00000519433.1
ENST00000517423.5 ENST00000220856.6 |
CCN4
|
cellular communication network factor 4 |
| chr1_-_17054015 | 0.04 |
ENST00000375499.8
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr4_-_64409444 | 0.04 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr20_+_48921701 | 0.04 |
ENST00000371917.5
|
ARFGEF2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr10_+_96130027 | 0.04 |
ENST00000478086.5
ENST00000624776.3 |
ZNF518A
|
zinc finger protein 518A |
| chr12_-_114406133 | 0.04 |
ENST00000405440.7
|
TBX5
|
T-box transcription factor 5 |
| chr9_+_110668854 | 0.04 |
ENST00000189978.10
ENST00000374440.7 |
MUSK
|
muscle associated receptor tyrosine kinase |
| chr11_-_790062 | 0.04 |
ENST00000330106.5
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr7_-_151736304 | 0.04 |
ENST00000492843.6
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr22_+_49960760 | 0.04 |
ENST00000360612.5
|
PIM3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr20_+_44715360 | 0.04 |
ENST00000190983.5
|
CCN5
|
cellular communication network factor 5 |
| chr11_-_132943671 | 0.04 |
ENST00000331898.11
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr2_+_218270392 | 0.04 |
ENST00000248451.7
ENST00000273077.9 |
PNKD
|
PNKD metallo-beta-lactamase domain containing |
| chr3_+_113532508 | 0.04 |
ENST00000264852.9
|
SIDT1
|
SID1 transmembrane family member 1 |
| chr11_+_78188871 | 0.04 |
ENST00000528910.5
ENST00000529308.6 |
USP35
|
ubiquitin specific peptidase 35 |
| chr9_+_113221528 | 0.04 |
ENST00000374212.5
|
SLC31A1
|
solute carrier family 31 member 1 |
| chr15_+_78149354 | 0.04 |
ENST00000558554.5
ENST00000557826.5 ENST00000561279.5 ENST00000299518.7 ENST00000559186.5 ENST00000560770.5 ENST00000559881.5 ENST00000559205.1 ENST00000629769.2 |
IDH3A
|
isocitrate dehydrogenase (NAD(+)) 3 catalytic subunit alpha |
| chr12_-_6606320 | 0.04 |
ENST00000642594.1
ENST00000644289.1 ENST00000645095.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr3_-_45958572 | 0.04 |
ENST00000433878.5
|
FYCO1
|
FYVE and coiled-coil domain autophagy adaptor 1 |
| chr10_+_96129707 | 0.04 |
ENST00000316045.9
|
ZNF518A
|
zinc finger protein 518A |
| chr12_-_16606795 | 0.04 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3 |
| chr1_-_201171545 | 0.04 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr7_+_45574358 | 0.04 |
ENST00000297323.12
|
ADCY1
|
adenylate cyclase 1 |
| chr19_-_5622768 | 0.04 |
ENST00000252542.9
|
SAFB2
|
scaffold attachment factor B2 |
| chr1_+_32292067 | 0.04 |
ENST00000373548.8
ENST00000428704.1 |
HDAC1
|
histone deacetylase 1 |
| chr10_+_45972482 | 0.04 |
ENST00000580018.4
|
TIMM23
|
translocase of inner mitochondrial membrane 23 |
| chr20_-_37527862 | 0.04 |
ENST00000373537.7
ENST00000445723.5 ENST00000414080.1 |
BLCAP
|
BLCAP apoptosis inducing factor |
| chr17_+_28744002 | 0.04 |
ENST00000618771.1
ENST00000262395.10 ENST00000422344.5 |
TRAF4
|
TNF receptor associated factor 4 |
| chr15_-_42491105 | 0.04 |
ENST00000565380.5
ENST00000564754.7 |
ZNF106
|
zinc finger protein 106 |
| chr16_-_87493013 | 0.04 |
ENST00000671377.2
|
ZCCHC14
|
zinc finger CCHC-type containing 14 |
| chr2_+_219434825 | 0.04 |
ENST00000312358.12
|
SPEG
|
striated muscle enriched protein kinase |
| chr9_+_126805003 | 0.04 |
ENST00000449886.5
ENST00000450858.1 ENST00000373464.5 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr4_-_73223082 | 0.03 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr6_+_106629560 | 0.03 |
ENST00000369046.8
|
QRSL1
|
glutaminyl-tRNA amidotransferase subunit QRSL1 |
| chr2_+_233778330 | 0.03 |
ENST00000389758.3
|
MROH2A
|
maestro heat like repeat family member 2A |
| chr3_-_113746185 | 0.03 |
ENST00000616174.1
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr4_-_121952014 | 0.03 |
ENST00000379645.8
|
TRPC3
|
transient receptor potential cation channel subfamily C member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.1 | 0.3 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.2 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.1 | 0.2 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 0.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0097017 | renal protein absorption(GO:0097017) |
| 0.0 | 0.1 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.6 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.3 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.6 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.3 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0046696 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0050567 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |