Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
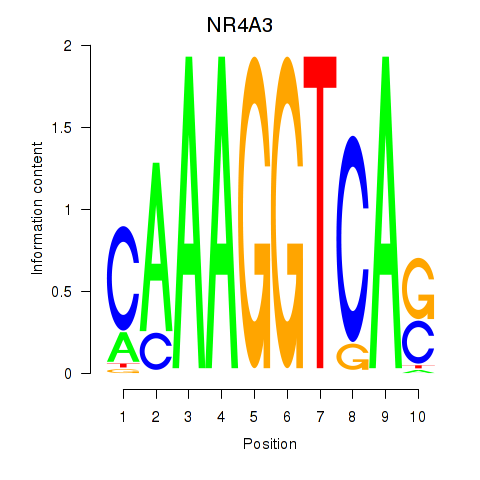
Results for NR4A3
Z-value: 0.65
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.18 | nuclear receptor subfamily 4 group A member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg38_v1_chr9_+_99821846_99821862 | 0.42 | 3.0e-01 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_33080445 | 0.44 |
ENST00000428835.5
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr10_-_104085847 | 0.36 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr17_-_41521719 | 0.34 |
ENST00000393976.6
|
KRT15
|
keratin 15 |
| chr2_-_31414694 | 0.33 |
ENST00000379416.4
|
XDH
|
xanthine dehydrogenase |
| chr22_+_20774092 | 0.30 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr15_-_82952683 | 0.30 |
ENST00000450735.7
ENST00000304231.12 |
HOMER2
|
homer scaffold protein 2 |
| chr6_-_11807045 | 0.29 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr6_+_33075952 | 0.28 |
ENST00000418931.7
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr1_-_175192911 | 0.28 |
ENST00000444639.5
|
KIAA0040
|
KIAA0040 |
| chr16_+_68645290 | 0.26 |
ENST00000264012.9
|
CDH3
|
cadherin 3 |
| chr14_-_21024092 | 0.26 |
ENST00000554398.5
|
NDRG2
|
NDRG family member 2 |
| chr17_+_39628496 | 0.26 |
ENST00000394265.5
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr15_+_43692886 | 0.26 |
ENST00000434505.5
ENST00000411750.5 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_+_43935807 | 0.25 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr14_-_21023954 | 0.24 |
ENST00000554094.5
|
NDRG2
|
NDRG family member 2 |
| chr1_+_152985231 | 0.24 |
ENST00000368762.1
|
SPRR1A
|
small proline rich protein 1A |
| chr3_-_151316795 | 0.22 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr10_-_114684612 | 0.22 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr18_+_11981488 | 0.21 |
ENST00000269159.8
|
IMPA2
|
inositol monophosphatase 2 |
| chr10_-_114684457 | 0.21 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr7_-_945799 | 0.21 |
ENST00000611167.4
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr5_-_74866958 | 0.21 |
ENST00000389156.9
|
FAM169A
|
family with sequence similarity 169 member A |
| chr15_+_59438149 | 0.21 |
ENST00000288228.10
ENST00000559628.5 ENST00000557914.5 ENST00000560474.5 |
FAM81A
|
family with sequence similarity 81 member A |
| chr20_+_59604527 | 0.20 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_+_148339452 | 0.20 |
ENST00000463592.3
|
CNTNAP2
|
contactin associated protein 2 |
| chr22_+_44676808 | 0.20 |
ENST00000624862.3
|
PRR5
|
proline rich 5 |
| chr18_+_11981548 | 0.19 |
ENST00000588927.5
|
IMPA2
|
inositol monophosphatase 2 |
| chr3_-_57079287 | 0.19 |
ENST00000338458.8
ENST00000468727.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr2_+_237859615 | 0.19 |
ENST00000409726.5
ENST00000254661.5 |
RAMP1
|
receptor activity modifying protein 1 |
| chr10_-_96271508 | 0.19 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr13_-_52011337 | 0.19 |
ENST00000400366.6
ENST00000400370.8 ENST00000634844.1 ENST00000673772.1 ENST00000418097.7 ENST00000242839.10 ENST00000344297.9 ENST00000448424.7 |
ATP7B
|
ATPase copper transporting beta |
| chr7_-_20217342 | 0.17 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr1_-_15585015 | 0.17 |
ENST00000375826.4
|
AGMAT
|
agmatinase |
| chr10_-_114404756 | 0.16 |
ENST00000369271.7
|
AFAP1L2
|
actin filament associated protein 1 like 2 |
| chr15_+_45129933 | 0.16 |
ENST00000321429.8
ENST00000389037.7 ENST00000558322.5 |
DUOX1
|
dual oxidase 1 |
| chr14_-_21098848 | 0.16 |
ENST00000556174.5
ENST00000554478.5 ENST00000553980.1 ENST00000421093.6 |
ZNF219
|
zinc finger protein 219 |
| chr2_+_219627394 | 0.16 |
ENST00000373760.6
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr12_+_112938284 | 0.16 |
ENST00000681346.1
|
OAS3
|
2'-5'-oligoadenylate synthetase 3 |
| chr19_+_35533436 | 0.16 |
ENST00000222286.9
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_+_67056805 | 0.16 |
ENST00000308831.7
|
RHOD
|
ras homolog family member D |
| chr1_+_159587817 | 0.16 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum |
| chr11_+_6845683 | 0.15 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr10_-_114404480 | 0.15 |
ENST00000419268.1
ENST00000304129.9 |
AFAP1L2
|
actin filament associated protein 1 like 2 |
| chr10_-_96271553 | 0.15 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr3_+_113897470 | 0.15 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr10_-_75235917 | 0.15 |
ENST00000469299.1
ENST00000372538.8 |
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr22_+_44677044 | 0.15 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chr22_+_44677077 | 0.15 |
ENST00000403581.5
|
PRR5
|
proline rich 5 |
| chr7_-_78771108 | 0.14 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_-_21098570 | 0.14 |
ENST00000360947.8
|
ZNF219
|
zinc finger protein 219 |
| chr19_-_38253238 | 0.14 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr6_-_43229451 | 0.14 |
ENST00000509253.5
ENST00000393987.2 ENST00000230431.11 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr7_+_69598465 | 0.14 |
ENST00000342771.10
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr1_+_6034677 | 0.14 |
ENST00000668559.1
|
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr11_+_67023085 | 0.14 |
ENST00000527043.6
|
SYT12
|
synaptotagmin 12 |
| chr8_+_85187650 | 0.14 |
ENST00000517476.5
ENST00000521429.5 |
E2F5
|
E2F transcription factor 5 |
| chr2_-_74440484 | 0.14 |
ENST00000305557.9
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr3_-_197573323 | 0.14 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr1_+_196888014 | 0.14 |
ENST00000367416.6
ENST00000608469.6 ENST00000251424.8 ENST00000367418.2 |
CFHR4
|
complement factor H related 4 |
| chr2_+_37231798 | 0.14 |
ENST00000439218.5
ENST00000432075.1 |
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr10_-_114526804 | 0.13 |
ENST00000369266.7
ENST00000369253.6 |
ABLIM1
|
actin binding LIM protein 1 |
| chr11_-_104164361 | 0.13 |
ENST00000302251.9
|
PDGFD
|
platelet derived growth factor D |
| chr17_-_7404039 | 0.13 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr7_+_69598292 | 0.13 |
ENST00000644939.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr7_+_118224654 | 0.13 |
ENST00000265224.9
ENST00000486422.1 ENST00000417525.5 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr2_+_237858874 | 0.13 |
ENST00000404910.6
|
RAMP1
|
receptor activity modifying protein 1 |
| chr12_+_108129276 | 0.13 |
ENST00000547525.6
|
WSCD2
|
WSC domain containing 2 |
| chr11_+_67056875 | 0.13 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr2_-_237590694 | 0.13 |
ENST00000264601.8
ENST00000411462.5 ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr7_-_81770122 | 0.13 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr1_+_1033987 | 0.12 |
ENST00000651234.1
ENST00000652369.1 |
AGRN
|
agrin |
| chr2_+_219627650 | 0.12 |
ENST00000317151.7
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr1_+_119507203 | 0.12 |
ENST00000369413.8
ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr2_-_237590660 | 0.12 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr1_+_24319342 | 0.12 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr3_-_185821092 | 0.12 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr14_-_103522696 | 0.12 |
ENST00000553878.5
ENST00000348956.7 ENST00000557530.1 |
CKB
|
creatine kinase B |
| chr8_+_135457442 | 0.12 |
ENST00000355849.10
|
KHDRBS3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr11_+_67266437 | 0.12 |
ENST00000308595.10
ENST00000526285.1 |
GRK2
|
G protein-coupled receptor kinase 2 |
| chr11_+_35176575 | 0.12 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr17_-_41118369 | 0.11 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr16_+_3654683 | 0.11 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr16_-_58295019 | 0.11 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr5_+_175658008 | 0.11 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr8_+_98944403 | 0.11 |
ENST00000457907.3
ENST00000523368.5 ENST00000297565.8 ENST00000435298.6 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr2_+_27014746 | 0.11 |
ENST00000648289.1
ENST00000458529.5 ENST00000402218.1 |
MAPRE3
|
microtubule associated protein RP/EB family member 3 |
| chr4_+_24795560 | 0.11 |
ENST00000382120.4
|
SOD3
|
superoxide dismutase 3 |
| chr11_+_10450627 | 0.11 |
ENST00000396554.7
ENST00000524866.5 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr18_+_58221535 | 0.11 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_+_24319511 | 0.11 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr1_+_26543106 | 0.11 |
ENST00000530003.5
|
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr12_+_112938523 | 0.11 |
ENST00000679483.1
ENST00000679493.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3 |
| chr19_-_42423100 | 0.11 |
ENST00000597001.1
|
LIPE
|
lipase E, hormone sensitive type |
| chr1_-_183590596 | 0.11 |
ENST00000418089.5
ENST00000413720.5 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr18_+_58149314 | 0.11 |
ENST00000435432.6
ENST00000357895.9 ENST00000586263.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr8_-_69833338 | 0.11 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr17_+_1742836 | 0.11 |
ENST00000324015.7
ENST00000450523.6 ENST00000453723.5 ENST00000453066.6 ENST00000382061.5 |
SERPINF2
|
serpin family F member 2 |
| chr4_-_7068033 | 0.11 |
ENST00000264954.5
|
GRPEL1
|
GrpE like 1, mitochondrial |
| chr14_-_21023318 | 0.11 |
ENST00000298684.9
ENST00000557169.5 ENST00000553563.5 |
NDRG2
|
NDRG family member 2 |
| chr16_+_57620077 | 0.11 |
ENST00000567835.5
ENST00000569372.5 ENST00000563548.5 ENST00000562003.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_144148027 | 0.11 |
ENST00000423230.6
|
MROH1
|
maestro heat like repeat family member 1 |
| chr4_-_149815826 | 0.11 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr16_-_58294976 | 0.10 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chr16_+_56657999 | 0.10 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr3_-_58666765 | 0.10 |
ENST00000358781.7
|
FAM3D
|
FAM3 metabolism regulating signaling molecule D |
| chr1_+_6034980 | 0.10 |
ENST00000378092.6
ENST00000472700.7 |
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr3_-_190449782 | 0.10 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chrX_+_103776493 | 0.10 |
ENST00000433491.5
ENST00000612423.4 ENST00000443502.5 |
PLP1
|
proteolipid protein 1 |
| chr19_-_48363914 | 0.10 |
ENST00000377431.6
ENST00000293261.8 |
TMEM143
|
transmembrane protein 143 |
| chr6_+_147508645 | 0.10 |
ENST00000367474.2
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr19_-_48364034 | 0.10 |
ENST00000435956.7
|
TMEM143
|
transmembrane protein 143 |
| chr11_-_117876892 | 0.10 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr3_+_113947901 | 0.10 |
ENST00000330212.7
ENST00000498275.5 |
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23 |
| chr14_-_106165730 | 0.10 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr1_-_1421302 | 0.10 |
ENST00000520296.5
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr6_-_43059367 | 0.10 |
ENST00000230413.9
ENST00000487429.1 ENST00000388752.8 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr12_+_53985138 | 0.10 |
ENST00000303460.5
|
HOXC10
|
homeobox C10 |
| chr5_+_36606355 | 0.10 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr11_-_117876719 | 0.10 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr17_+_41105332 | 0.10 |
ENST00000391415.1
ENST00000617453.1 |
KRTAP4-9
|
keratin associated protein 4-9 |
| chr17_-_6713359 | 0.10 |
ENST00000381074.8
ENST00000433363.7 ENST00000293800.10 ENST00000572352.5 ENST00000573648.5 |
SLC13A5
|
solute carrier family 13 member 5 |
| chr18_+_58862904 | 0.09 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532 |
| chr19_-_43639788 | 0.09 |
ENST00000222374.3
|
CADM4
|
cell adhesion molecule 4 |
| chr11_-_78023214 | 0.09 |
ENST00000353172.6
|
KCTD14
|
potassium channel tetramerization domain containing 14 |
| chr11_+_10455292 | 0.09 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr17_-_42112674 | 0.09 |
ENST00000251642.8
ENST00000591220.5 |
DHX58
|
DExH-box helicase 58 |
| chr19_+_50770464 | 0.09 |
ENST00000270590.4
|
GPR32
|
G protein-coupled receptor 32 |
| chr11_-_8810635 | 0.09 |
ENST00000527510.5
ENST00000528527.5 ENST00000313726.11 ENST00000528523.5 |
DENND2B
|
DENN domain containing 2B |
| chr5_+_36166556 | 0.09 |
ENST00000677886.1
|
SKP2
|
S-phase kinase associated protein 2 |
| chr2_+_219442023 | 0.09 |
ENST00000431523.5
ENST00000396698.5 |
SPEG
|
striated muscle enriched protein kinase |
| chr1_+_46303646 | 0.09 |
ENST00000311672.10
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr12_+_69825221 | 0.09 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr1_-_1421248 | 0.09 |
ENST00000442470.1
ENST00000537107.6 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr7_-_37916807 | 0.09 |
ENST00000436072.7
|
SFRP4
|
secreted frizzled related protein 4 |
| chr4_-_40515967 | 0.09 |
ENST00000381795.10
|
RBM47
|
RNA binding motif protein 47 |
| chr16_-_20697680 | 0.09 |
ENST00000520010.6
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr17_+_4771878 | 0.09 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr12_+_53423849 | 0.09 |
ENST00000257863.9
ENST00000550311.5 ENST00000379791.7 |
AMHR2
|
anti-Mullerian hormone receptor type 2 |
| chr16_+_69105636 | 0.09 |
ENST00000569188.6
|
HAS3
|
hyaluronan synthase 3 |
| chr19_-_633500 | 0.09 |
ENST00000588649.7
|
POLRMT
|
RNA polymerase mitochondrial |
| chr19_+_17305801 | 0.09 |
ENST00000602206.1
ENST00000252602.2 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr9_+_128322540 | 0.09 |
ENST00000609948.1
ENST00000608951.5 |
COQ4
|
coenzyme Q4 |
| chr2_+_233778330 | 0.08 |
ENST00000389758.3
|
MROH2A
|
maestro heat like repeat family member 2A |
| chr4_+_41612892 | 0.08 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr22_+_46150590 | 0.08 |
ENST00000262735.9
ENST00000420804.5 |
PPARA
|
peroxisome proliferator activated receptor alpha |
| chr2_+_176157293 | 0.08 |
ENST00000683222.1
|
HOXD3
|
homeobox D3 |
| chr3_+_158571153 | 0.08 |
ENST00000491767.6
ENST00000618075.4 |
MLF1
|
myeloid leukemia factor 1 |
| chr3_+_113948004 | 0.08 |
ENST00000638807.2
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23 |
| chr11_-_104164126 | 0.08 |
ENST00000393158.7
|
PDGFD
|
platelet derived growth factor D |
| chr4_-_75902444 | 0.08 |
ENST00000286719.12
|
PPEF2
|
protein phosphatase with EF-hand domain 2 |
| chr15_+_58431985 | 0.08 |
ENST00000433326.2
ENST00000299022.10 |
LIPC
|
lipase C, hepatic type |
| chr17_-_79950828 | 0.08 |
ENST00000572862.5
ENST00000573782.5 ENST00000574427.1 ENST00000570373.5 ENST00000340848.11 ENST00000576768.5 |
TBC1D16
|
TBC1 domain family member 16 |
| chr1_-_46303589 | 0.08 |
ENST00000617190.5
|
LRRC41
|
leucine rich repeat containing 41 |
| chr15_-_82647960 | 0.08 |
ENST00000615198.4
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr1_+_171257930 | 0.08 |
ENST00000354841.4
|
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr16_+_57619942 | 0.08 |
ENST00000568908.5
ENST00000568909.5 ENST00000566778.5 ENST00000561988.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr19_-_11529094 | 0.08 |
ENST00000588998.5
ENST00000586149.1 |
ECSIT
|
ECSIT signaling integrator |
| chrX_-_54998530 | 0.08 |
ENST00000545676.5
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_+_44874606 | 0.08 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr16_-_30526758 | 0.08 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chr17_-_1649854 | 0.08 |
ENST00000301336.7
|
RILP
|
Rab interacting lysosomal protein |
| chr3_+_184337591 | 0.08 |
ENST00000383847.7
|
FAM131A
|
family with sequence similarity 131 member A |
| chr19_+_1205761 | 0.08 |
ENST00000326873.12
ENST00000586243.5 |
STK11
|
serine/threonine kinase 11 |
| chr10_-_49762335 | 0.08 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr11_-_8932944 | 0.08 |
ENST00000326053.10
ENST00000525780.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr3_+_158571171 | 0.08 |
ENST00000484955.5
ENST00000359117.9 ENST00000619577.5 ENST00000471745.5 ENST00000477042.6 ENST00000650753.1 ENST00000651984.1 ENST00000355893.11 ENST00000466246.7 ENST00000469452.5 ENST00000482628.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr5_+_1801387 | 0.08 |
ENST00000274137.10
ENST00000469176.1 |
NDUFS6
|
NADH:ubiquinone oxidoreductase subunit S6 |
| chr1_+_171248471 | 0.08 |
ENST00000402921.6
ENST00000617670.6 ENST00000367750.7 |
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr19_-_42879635 | 0.08 |
ENST00000595356.5
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr15_+_90184912 | 0.07 |
ENST00000561085.1
ENST00000332496.10 |
SEMA4B
|
semaphorin 4B |
| chr1_+_23791138 | 0.07 |
ENST00000374514.8
ENST00000420982.5 ENST00000374505.6 |
LYPLA2
|
lysophospholipase 2 |
| chr7_-_111562455 | 0.07 |
ENST00000452895.5
ENST00000405709.7 ENST00000452753.1 ENST00000331762.7 |
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr19_-_11529116 | 0.07 |
ENST00000592312.5
ENST00000590480.1 ENST00000585318.5 ENST00000270517.12 ENST00000252440.11 ENST00000417981.6 |
ECSIT
|
ECSIT signaling integrator |
| chr12_+_69825273 | 0.07 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr7_-_78771265 | 0.07 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_+_132490140 | 0.07 |
ENST00000680143.1
|
FBRSL1
|
fibrosin like 1 |
| chr19_-_1174227 | 0.07 |
ENST00000587024.5
ENST00000361757.8 |
SBNO2
|
strawberry notch homolog 2 |
| chr8_+_144095054 | 0.07 |
ENST00000318911.5
|
CYC1
|
cytochrome c1 |
| chr1_+_162381703 | 0.07 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr10_+_79347491 | 0.07 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chrX_+_66164340 | 0.07 |
ENST00000441993.7
ENST00000419594.6 ENST00000425114.2 |
HEPH
|
hephaestin |
| chr4_+_41612702 | 0.07 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr16_+_85908988 | 0.07 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr11_-_67469210 | 0.07 |
ENST00000393877.3
ENST00000308022.7 |
TMEM134
|
transmembrane protein 134 |
| chrX_+_23783163 | 0.07 |
ENST00000379254.5
ENST00000379270.5 ENST00000683890.1 |
SAT1
ENSG00000288706.1
|
spermidine/spermine N1-acetyltransferase 1 novel protein |
| chr19_-_2328573 | 0.07 |
ENST00000587502.2
ENST00000252622.15 ENST00000585409.2 |
LSM7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_+_132673983 | 0.07 |
ENST00000622422.1
ENST00000231449.7 ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr22_+_20117424 | 0.07 |
ENST00000402752.5
|
RANBP1
|
RAN binding protein 1 |
| chr16_-_67416420 | 0.07 |
ENST00000348579.6
ENST00000565726.3 |
ZDHHC1
|
zinc finger DHHC-type containing 1 |
| chr17_-_2711736 | 0.07 |
ENST00000651024.2
ENST00000576885.5 ENST00000574426.7 |
CLUH
|
clustered mitochondria homolog |
| chr6_+_18155399 | 0.07 |
ENST00000650836.2
ENST00000449850.2 ENST00000297792.9 |
KDM1B
|
lysine demethylase 1B |
| chr19_-_29213110 | 0.07 |
ENST00000304863.6
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr2_+_106487349 | 0.06 |
ENST00000643224.2
ENST00000416057.2 |
CD8B2
|
CD8b2 molecule |
| chr19_-_57935353 | 0.06 |
ENST00000595569.1
ENST00000599852.1 ENST00000425570.7 ENST00000601593.5 ENST00000396147.6 |
ZNF418
|
zinc finger protein 418 |
| chr10_+_116621306 | 0.06 |
ENST00000611850.4
ENST00000591655.3 |
PNLIPRP2
|
pancreatic lipase related protein 2 (gene/pseudogene) |
| chr12_-_95116967 | 0.06 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chrX_+_47218670 | 0.06 |
ENST00000357227.9
ENST00000519758.5 ENST00000520893.5 ENST00000622098.4 ENST00000517426.5 |
CDK16
|
cyclin dependent kinase 16 |
| chr3_+_158571215 | 0.06 |
ENST00000498592.6
ENST00000478894.7 |
MLF1
|
myeloid leukemia factor 1 |
| chr1_+_212565334 | 0.06 |
ENST00000366981.8
ENST00000366987.6 |
ATF3
|
activating transcription factor 3 |
| chr2_+_88691647 | 0.06 |
ENST00000283646.5
|
RPIA
|
ribose 5-phosphate isomerase A |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.3 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.4 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.2 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 0.2 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.3 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.0 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.3 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0060503 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.2 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.2 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.0 | GO:0018013 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.0 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.0 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |