Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
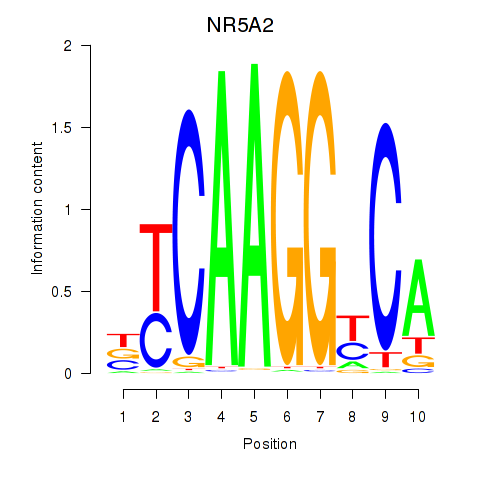
Results for NR5A2
Z-value: 0.84
Transcription factors associated with NR5A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR5A2
|
ENSG00000116833.14 | nuclear receptor subfamily 5 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR5A2 | hg38_v1_chr1_+_200027702_200027716 | -0.71 | 4.7e-02 | Click! |
Activity profile of NR5A2 motif
Sorted Z-values of NR5A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_59577463 | 1.34 |
ENST00000359926.7
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr10_+_116427839 | 0.86 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr20_+_59604527 | 0.82 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_+_219627394 | 0.77 |
ENST00000373760.6
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr15_+_43593054 | 0.71 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr15_+_43692886 | 0.69 |
ENST00000434505.5
ENST00000411750.5 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr6_-_136526177 | 0.56 |
ENST00000617204.4
|
MAP7
|
microtubule associated protein 7 |
| chr6_-_136525961 | 0.45 |
ENST00000438100.6
|
MAP7
|
microtubule associated protein 7 |
| chr11_-_72785932 | 0.45 |
ENST00000539138.1
ENST00000542989.5 |
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr22_+_39994926 | 0.42 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F |
| chr3_-_49903863 | 0.41 |
ENST00000296474.8
ENST00000621387.4 |
MST1R
|
macrophage stimulating 1 receptor |
| chr2_+_219627565 | 0.41 |
ENST00000273063.10
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr6_+_30888672 | 0.39 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_219627650 | 0.39 |
ENST00000317151.7
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr2_+_219627622 | 0.38 |
ENST00000358055.8
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr12_-_106138946 | 0.38 |
ENST00000261402.7
|
NUAK1
|
NUAK family kinase 1 |
| chr11_+_10450627 | 0.38 |
ENST00000396554.7
ENST00000524866.5 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr15_-_82952683 | 0.37 |
ENST00000450735.7
ENST00000304231.12 |
HOMER2
|
homer scaffold protein 2 |
| chr1_+_152908538 | 0.35 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr2_-_237590660 | 0.34 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr9_-_114806031 | 0.34 |
ENST00000374045.5
|
TNFSF15
|
TNF superfamily member 15 |
| chr15_+_45129933 | 0.33 |
ENST00000321429.8
ENST00000389037.7 ENST00000558322.5 |
DUOX1
|
dual oxidase 1 |
| chr15_-_81324130 | 0.33 |
ENST00000302824.7
|
STARD5
|
StAR related lipid transfer domain containing 5 |
| chr19_+_48270079 | 0.32 |
ENST00000595607.6
|
ZNF114
|
zinc finger protein 114 |
| chr8_+_22053543 | 0.31 |
ENST00000519850.5
ENST00000381470.7 |
DMTN
|
dematin actin binding protein |
| chr17_-_29176752 | 0.30 |
ENST00000533112.5
|
MYO18A
|
myosin XVIIIA |
| chr5_+_52989314 | 0.30 |
ENST00000296585.10
|
ITGA2
|
integrin subunit alpha 2 |
| chr11_+_59713403 | 0.30 |
ENST00000641815.1
|
STX3
|
syntaxin 3 |
| chr1_-_11654422 | 0.30 |
ENST00000354287.5
|
FBXO2
|
F-box protein 2 |
| chr19_+_45340736 | 0.29 |
ENST00000391946.7
|
KLC3
|
kinesin light chain 3 |
| chr19_+_45340760 | 0.29 |
ENST00000585434.5
|
KLC3
|
kinesin light chain 3 |
| chr2_-_31138429 | 0.29 |
ENST00000349752.10
|
GALNT14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr19_+_14941489 | 0.28 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor family 7 subfamily C member 2 |
| chr10_-_75235917 | 0.28 |
ENST00000469299.1
ENST00000372538.8 |
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr14_-_23155302 | 0.28 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr12_-_56488350 | 0.27 |
ENST00000623608.3
ENST00000610413.4 |
GLS2
|
glutaminase 2 |
| chr2_-_237590694 | 0.27 |
ENST00000264601.8
ENST00000411462.5 ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr12_+_132490140 | 0.26 |
ENST00000680143.1
|
FBRSL1
|
fibrosin like 1 |
| chr10_-_86969178 | 0.26 |
ENST00000440490.1
ENST00000609457.5 |
ADIRF-AS1
MMRN2
|
ADIRF antisense RNA 1 multimerin 2 |
| chr3_+_113947901 | 0.25 |
ENST00000330212.7
ENST00000498275.5 |
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23 |
| chr1_+_15756659 | 0.25 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr12_+_100794769 | 0.25 |
ENST00000392977.8
ENST00000546991.1 ENST00000392979.7 |
ANO4
|
anoctamin 4 |
| chr19_+_2249317 | 0.25 |
ENST00000221496.5
|
AMH
|
anti-Mullerian hormone |
| chr15_-_51243011 | 0.25 |
ENST00000405913.7
ENST00000559878.5 |
CYP19A1
|
cytochrome P450 family 19 subfamily A member 1 |
| chr3_+_186996444 | 0.25 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr3_-_121660892 | 0.24 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr12_+_43836043 | 0.24 |
ENST00000266534.8
ENST00000551577.5 |
TMEM117
|
transmembrane protein 117 |
| chr7_+_130266847 | 0.24 |
ENST00000222481.9
|
CPA2
|
carboxypeptidase A2 |
| chr17_-_40501615 | 0.24 |
ENST00000254051.11
|
TNS4
|
tensin 4 |
| chr22_+_48693204 | 0.23 |
ENST00000406880.1
|
TAFA5
|
TAFA chemokine like family member 5 |
| chr22_+_44677044 | 0.23 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chr1_+_32765667 | 0.23 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr22_+_44677077 | 0.23 |
ENST00000403581.5
|
PRR5
|
proline rich 5 |
| chr22_+_44676808 | 0.23 |
ENST00000624862.3
|
PRR5
|
proline rich 5 |
| chr3_+_113948004 | 0.23 |
ENST00000638807.2
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23 |
| chr19_+_45340774 | 0.23 |
ENST00000589837.5
|
KLC3
|
kinesin light chain 3 |
| chr21_+_42219123 | 0.22 |
ENST00000398449.8
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr2_+_46297397 | 0.22 |
ENST00000263734.5
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr4_-_170026371 | 0.22 |
ENST00000361618.4
ENST00000506764.1 |
MFAP3L
|
microfibril associated protein 3 like |
| chr17_+_32487686 | 0.22 |
ENST00000584792.5
|
CDK5R1
|
cyclin dependent kinase 5 regulatory subunit 1 |
| chr5_-_60844185 | 0.21 |
ENST00000505959.5
|
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr16_+_67431112 | 0.21 |
ENST00000326152.6
|
HSD11B2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr21_+_42219111 | 0.21 |
ENST00000450121.5
ENST00000361802.6 |
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr10_+_79347491 | 0.19 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr11_-_72642450 | 0.19 |
ENST00000444035.6
ENST00000544570.5 |
PDE2A
|
phosphodiesterase 2A |
| chr4_-_170026333 | 0.19 |
ENST00000504999.1
|
MFAP3L
|
microfibril associated protein 3 like |
| chr11_-_72642407 | 0.19 |
ENST00000376450.7
|
PDE2A
|
phosphodiesterase 2A |
| chr15_+_90184912 | 0.19 |
ENST00000561085.1
ENST00000332496.10 |
SEMA4B
|
semaphorin 4B |
| chr21_+_38272291 | 0.18 |
ENST00000438657.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr16_-_84239750 | 0.18 |
ENST00000568181.1
|
KCNG4
|
potassium voltage-gated channel modifier subfamily G member 4 |
| chr22_-_39244969 | 0.18 |
ENST00000331163.11
|
PDGFB
|
platelet derived growth factor subunit B |
| chr10_-_49762335 | 0.17 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr2_-_236507428 | 0.17 |
ENST00000409907.8
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr8_-_130016414 | 0.17 |
ENST00000401979.6
ENST00000517654.5 ENST00000522361.1 ENST00000518167.5 |
CYRIB
|
CYFIP related Rac1 interactor B |
| chr1_+_207053229 | 0.17 |
ENST00000367080.8
ENST00000367079.3 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr2_+_233195433 | 0.17 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr17_+_7705193 | 0.17 |
ENST00000226091.3
|
EFNB3
|
ephrin B3 |
| chr6_-_27132750 | 0.16 |
ENST00000607124.1
ENST00000339812.3 |
H2BC11
|
H2B clustered histone 11 |
| chr14_+_93927259 | 0.16 |
ENST00000556222.1
ENST00000554404.1 |
FAM181A
|
family with sequence similarity 181 member A |
| chr2_-_236507515 | 0.16 |
ENST00000309507.9
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr19_-_42877988 | 0.16 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr6_-_143511682 | 0.16 |
ENST00000002165.11
|
FUCA2
|
alpha-L-fucosidase 2 |
| chr17_+_7439504 | 0.16 |
ENST00000575331.1
ENST00000293829.9 |
ENSG00000272884.1
FGF11
|
novel transcript fibroblast growth factor 11 |
| chr5_-_16616972 | 0.16 |
ENST00000682564.1
ENST00000306320.10 ENST00000682229.1 |
RETREG1
|
reticulophagy regulator 1 |
| chr5_-_16617085 | 0.16 |
ENST00000684521.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr4_-_75630473 | 0.15 |
ENST00000307465.9
|
CDKL2
|
cyclin dependent kinase like 2 |
| chr6_+_116461364 | 0.15 |
ENST00000368606.7
ENST00000368605.3 |
CALHM6
|
calcium homeostasis modulator family member 6 |
| chr1_+_26543106 | 0.15 |
ENST00000530003.5
|
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr1_-_85048437 | 0.15 |
ENST00000341115.8
ENST00000370587.5 ENST00000370589.7 |
MCOLN3
|
mucolipin TRP cation channel 3 |
| chrX_+_49058966 | 0.15 |
ENST00000496529.6
ENST00000606812.5 ENST00000603986.6 ENST00000536628.3 |
CCDC120
|
coiled-coil domain containing 120 |
| chr17_+_50508384 | 0.15 |
ENST00000436259.6
ENST00000652471.1 ENST00000323776.11 ENST00000419930.2 |
MYCBPAP
|
MYCBP associated protein |
| chr16_-_4538469 | 0.15 |
ENST00000588381.1
ENST00000563332.6 |
CDIP1
|
cell death inducing p53 target 1 |
| chr11_+_66291887 | 0.14 |
ENST00000327259.5
|
TMEM151A
|
transmembrane protein 151A |
| chr19_-_51026593 | 0.14 |
ENST00000600362.5
ENST00000453757.8 ENST00000601671.1 |
KLK11
|
kallikrein related peptidase 11 |
| chr1_+_42153399 | 0.14 |
ENST00000372581.2
|
GUCA2B
|
guanylate cyclase activator 2B |
| chr1_-_149886652 | 0.14 |
ENST00000369155.3
|
H2BC21
|
H2B clustered histone 21 |
| chr22_+_26483851 | 0.14 |
ENST00000215917.11
|
SRRD
|
SRR1 domain containing |
| chr12_-_7092422 | 0.14 |
ENST00000543835.5
ENST00000647956.2 ENST00000535233.6 |
C1R
|
complement C1r |
| chrX_-_49200174 | 0.14 |
ENST00000472598.5
ENST00000263233.9 ENST00000479808.5 |
SYP
|
synaptophysin |
| chr15_+_64911869 | 0.14 |
ENST00000319580.13
ENST00000496660.5 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr19_+_41797147 | 0.14 |
ENST00000596544.1
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr3_-_122793772 | 0.13 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr11_-_124897797 | 0.13 |
ENST00000306534.8
ENST00000533054.5 |
ROBO4
|
roundabout guidance receptor 4 |
| chr17_-_4560564 | 0.13 |
ENST00000574584.1
ENST00000381550.8 ENST00000301395.7 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr9_-_129178247 | 0.13 |
ENST00000372491.4
|
IER5L
|
immediate early response 5 like |
| chr21_+_38272410 | 0.13 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr16_-_66918839 | 0.13 |
ENST00000565235.2
ENST00000568632.5 ENST00000565796.5 |
CDH16
|
cadherin 16 |
| chr5_+_66958870 | 0.13 |
ENST00000405643.5
ENST00000407621.1 ENST00000432426.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_147225309 | 0.13 |
ENST00000369272.7
ENST00000254090.9 ENST00000441068.6 |
FMO5
|
flavin containing dimethylaniline monoxygenase 5 |
| chr10_+_116324440 | 0.13 |
ENST00000333254.4
|
CCDC172
|
coiled-coil domain containing 172 |
| chr10_+_72893734 | 0.13 |
ENST00000334011.10
|
OIT3
|
oncoprotein induced transcript 3 |
| chr21_+_38272250 | 0.13 |
ENST00000398932.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr16_-_66918876 | 0.13 |
ENST00000570262.5
ENST00000299752.9 ENST00000394055.7 |
CDH16
|
cadherin 16 |
| chr1_+_32741779 | 0.13 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr6_+_89080739 | 0.13 |
ENST00000369472.1
ENST00000336032.4 |
PNRC1
|
proline rich nuclear receptor coactivator 1 |
| chr13_-_110561668 | 0.13 |
ENST00000267328.5
|
RAB20
|
RAB20, member RAS oncogene family |
| chr17_+_7435416 | 0.13 |
ENST00000323206.2
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr12_+_6385119 | 0.13 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor |
| chr3_-_10708007 | 0.13 |
ENST00000646379.1
|
ATP2B2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr4_-_170027209 | 0.13 |
ENST00000393702.7
|
MFAP3L
|
microfibril associated protein 3 like |
| chr7_+_75398142 | 0.13 |
ENST00000447409.6
|
TRIM73
|
tripartite motif containing 73 |
| chr10_+_79347460 | 0.12 |
ENST00000225174.8
|
PPIF
|
peptidylprolyl isomerase F |
| chr15_+_59438149 | 0.12 |
ENST00000288228.10
ENST00000559628.5 ENST00000557914.5 ENST00000560474.5 |
FAM81A
|
family with sequence similarity 81 member A |
| chr6_+_27815010 | 0.12 |
ENST00000621112.2
|
H2BC14
|
H2B clustered histone 14 |
| chr10_+_23439060 | 0.12 |
ENST00000376495.5
|
OTUD1
|
OTU deubiquitinase 1 |
| chr12_+_56338873 | 0.12 |
ENST00000228534.6
|
IL23A
|
interleukin 23 subunit alpha |
| chr7_+_48089257 | 0.12 |
ENST00000436673.5
ENST00000395564.9 |
UPP1
|
uridine phosphorylase 1 |
| chr17_+_1742836 | 0.12 |
ENST00000324015.7
ENST00000450523.6 ENST00000453723.5 ENST00000453066.6 ENST00000382061.5 |
SERPINF2
|
serpin family F member 2 |
| chr17_-_40100569 | 0.12 |
ENST00000246672.4
|
NR1D1
|
nuclear receptor subfamily 1 group D member 1 |
| chr4_+_73436198 | 0.12 |
ENST00000395792.7
|
AFP
|
alpha fetoprotein |
| chr8_-_130016395 | 0.12 |
ENST00000523509.5
|
CYRIB
|
CYFIP related Rac1 interactor B |
| chr14_-_103522696 | 0.12 |
ENST00000553878.5
ENST00000348956.7 ENST00000557530.1 |
CKB
|
creatine kinase B |
| chr2_-_175181663 | 0.12 |
ENST00000392541.3
ENST00000284727.9 ENST00000409194.5 |
ATP5MC3
|
ATP synthase membrane subunit c locus 3 |
| chr8_+_124539097 | 0.12 |
ENST00000606244.2
ENST00000276689.8 ENST00000518008.5 ENST00000517367.1 |
NDUFB9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr19_-_17026795 | 0.12 |
ENST00000443236.7
|
CPAMD8
|
C3 and PZP like alpha-2-macroglobulin domain containing 8 |
| chr7_-_72966953 | 0.12 |
ENST00000395244.5
|
TRIM74
|
tripartite motif containing 74 |
| chr4_+_73436244 | 0.12 |
ENST00000226359.2
|
AFP
|
alpha fetoprotein |
| chr1_+_115976488 | 0.12 |
ENST00000369503.9
|
SLC22A15
|
solute carrier family 22 member 15 |
| chr17_-_75509880 | 0.11 |
ENST00000433559.6
|
CASKIN2
|
CASK interacting protein 2 |
| chr22_-_29315656 | 0.11 |
ENST00000401450.3
ENST00000216101.7 |
RASL10A
|
RAS like family 10 member A |
| chr1_+_43979179 | 0.11 |
ENST00000434555.7
ENST00000372324.6 ENST00000481924.2 |
B4GALT2
|
beta-1,4-galactosyltransferase 2 |
| chr4_-_10457385 | 0.11 |
ENST00000507515.1
ENST00000326756.4 |
ZNF518B
|
zinc finger protein 518B |
| chr8_+_102551583 | 0.11 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr18_-_31162849 | 0.11 |
ENST00000257197.7
ENST00000257198.6 |
DSC1
|
desmocollin 1 |
| chr2_+_219442023 | 0.11 |
ENST00000431523.5
ENST00000396698.5 |
SPEG
|
striated muscle enriched protein kinase |
| chr20_+_1895365 | 0.11 |
ENST00000358771.5
|
SIRPA
|
signal regulatory protein alpha |
| chr17_-_2711736 | 0.11 |
ENST00000651024.2
ENST00000576885.5 ENST00000574426.7 |
CLUH
|
clustered mitochondria homolog |
| chr1_-_149842736 | 0.11 |
ENST00000369159.2
|
H2AC18
|
H2A clustered histone 18 |
| chr1_+_19596960 | 0.11 |
ENST00000617872.4
ENST00000322753.7 ENST00000602662.1 |
MICOS10
NBL1
|
mitochondrial contact site and cristae organizing system subunit 10 NBL1, DAN family BMP antagonist |
| chr6_+_143864458 | 0.11 |
ENST00000237275.9
|
ZC2HC1B
|
zinc finger C2HC-type containing 1B |
| chr14_-_21023318 | 0.11 |
ENST00000298684.9
ENST00000557169.5 ENST00000553563.5 |
NDRG2
|
NDRG family member 2 |
| chr6_-_42194947 | 0.11 |
ENST00000230361.4
|
GUCA1B
|
guanylate cyclase activator 1B |
| chr3_-_108953870 | 0.11 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr11_+_7576408 | 0.11 |
ENST00000533792.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr3_+_49689531 | 0.11 |
ENST00000432042.5
ENST00000454491.5 ENST00000327697.11 |
RNF123
|
ring finger protein 123 |
| chr2_-_241637045 | 0.11 |
ENST00000407315.6
|
THAP4
|
THAP domain containing 4 |
| chr12_+_57583101 | 0.11 |
ENST00000674858.1
ENST00000675433.1 ENST00000674980.1 |
KIF5A
|
kinesin family member 5A |
| chr14_-_21022817 | 0.11 |
ENST00000554104.5
|
NDRG2
|
NDRG family member 2 |
| chr1_+_32251239 | 0.11 |
ENST00000373564.7
ENST00000482949.5 ENST00000336890.10 ENST00000495610.6 |
LCK
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr8_-_130016622 | 0.10 |
ENST00000518283.5
ENST00000519110.5 |
CYRIB
|
CYFIP related Rac1 interactor B |
| chr2_+_176269406 | 0.10 |
ENST00000249442.11
ENST00000443241.5 |
MTX2
|
metaxin 2 |
| chr4_-_73998669 | 0.10 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5 |
| chr17_-_29589536 | 0.10 |
ENST00000394869.7
|
GIT1
|
GIT ArfGAP 1 |
| chr19_-_17245889 | 0.10 |
ENST00000291442.4
|
NR2F6
|
nuclear receptor subfamily 2 group F member 6 |
| chr22_+_46150590 | 0.10 |
ENST00000262735.9
ENST00000420804.5 |
PPARA
|
peroxisome proliferator activated receptor alpha |
| chr11_-_95232514 | 0.10 |
ENST00000634898.1
ENST00000542176.1 ENST00000278499.6 |
SESN3
|
sestrin 3 |
| chr14_-_91060113 | 0.10 |
ENST00000536315.6
|
RPS6KA5
|
ribosomal protein S6 kinase A5 |
| chr2_+_237966955 | 0.10 |
ENST00000414443.5
ENST00000448502.5 ENST00000416292.5 ENST00000409633.5 ENST00000272930.9 ENST00000409953.5 ENST00000409332.5 |
UBE2F
|
ubiquitin conjugating enzyme E2 F (putative) |
| chr17_-_29589606 | 0.10 |
ENST00000225394.8
|
GIT1
|
GIT ArfGAP 1 |
| chr8_-_58500158 | 0.10 |
ENST00000301645.4
|
CYP7A1
|
cytochrome P450 family 7 subfamily A member 1 |
| chr7_+_151085858 | 0.10 |
ENST00000463381.5
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr19_+_44748673 | 0.10 |
ENST00000164227.10
|
BCL3
|
BCL3 transcription coactivator |
| chr14_-_103521342 | 0.10 |
ENST00000553610.5
|
CKB
|
creatine kinase B |
| chr20_-_45631235 | 0.09 |
ENST00000326000.2
|
WFDC9
|
WAP four-disulfide core domain 9 |
| chr11_+_26332117 | 0.09 |
ENST00000531646.1
ENST00000256737.8 |
ANO3
|
anoctamin 3 |
| chr8_+_144095054 | 0.09 |
ENST00000318911.5
|
CYC1
|
cytochrome c1 |
| chr21_+_41167774 | 0.09 |
ENST00000328735.10
ENST00000347667.5 |
BACE2
|
beta-secretase 2 |
| chr5_+_141051374 | 0.09 |
ENST00000306549.6
|
PCDHB1
|
protocadherin beta 1 |
| chr5_+_141637398 | 0.09 |
ENST00000518856.1
|
RELL2
|
RELT like 2 |
| chr1_+_110210272 | 0.09 |
ENST00000438661.3
|
KCNC4
|
potassium voltage-gated channel subfamily C member 4 |
| chr7_+_55964577 | 0.09 |
ENST00000446778.5
ENST00000322090.8 |
NIPSNAP2
|
nipsnap homolog 2 |
| chr16_+_641792 | 0.09 |
ENST00000307650.9
ENST00000629534.2 |
MCRIP2
|
MAPK regulated corepressor interacting protein 2 |
| chr9_-_127916978 | 0.09 |
ENST00000361444.3
ENST00000335791.10 |
ST6GALNAC4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chrX_+_48786578 | 0.09 |
ENST00000376670.9
|
GATA1
|
GATA binding protein 1 |
| chr12_-_109833373 | 0.09 |
ENST00000261740.7
|
TRPV4
|
transient receptor potential cation channel subfamily V member 4 |
| chrX_+_155026835 | 0.09 |
ENST00000369498.8
|
FUNDC2
|
FUN14 domain containing 2 |
| chr17_-_58007217 | 0.09 |
ENST00000258962.5
ENST00000582730.6 ENST00000584773.5 ENST00000585096.1 |
SRSF1
|
serine and arginine rich splicing factor 1 |
| chr16_-_58295019 | 0.09 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr20_+_43916142 | 0.09 |
ENST00000423191.6
ENST00000372999.5 |
TOX2
|
TOX high mobility group box family member 2 |
| chr20_+_54475584 | 0.09 |
ENST00000262593.10
|
DOK5
|
docking protein 5 |
| chr7_-_945799 | 0.09 |
ENST00000611167.4
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr17_-_40937445 | 0.09 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr5_-_74866958 | 0.09 |
ENST00000389156.9
|
FAM169A
|
family with sequence similarity 169 member A |
| chr17_-_8087709 | 0.09 |
ENST00000647874.1
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chr14_+_92513766 | 0.09 |
ENST00000216487.12
ENST00000620541.4 ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr18_+_44680875 | 0.09 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr3_+_4493340 | 0.09 |
ENST00000357086.10
ENST00000354582.12 ENST00000649015.2 ENST00000467056.6 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr18_+_44680093 | 0.09 |
ENST00000426838.8
ENST00000677068.1 |
SETBP1
|
SET binding protein 1 |
| chr10_-_123008784 | 0.09 |
ENST00000368886.10
|
IKZF5
|
IKAROS family zinc finger 5 |
| chr11_-_1762403 | 0.09 |
ENST00000367196.4
|
CTSD
|
cathepsin D |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR5A2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.4 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 0.3 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.4 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.2 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.2 | GO:0072209 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 0.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.4 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.1 | GO:0070859 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.6 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.3 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.0 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0060406 | maternal aggressive behavior(GO:0002125) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.7 | GO:0008088 | axo-dendritic transport(GO:0008088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 0.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.7 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0010465 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0008443 | 6-phosphofructokinase activity(GO:0003872) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0050733 | DNA topoisomerase binding(GO:0044547) RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |