Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Z-value: 1.22
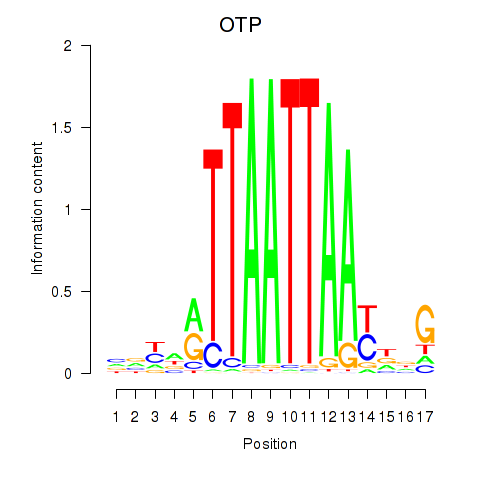
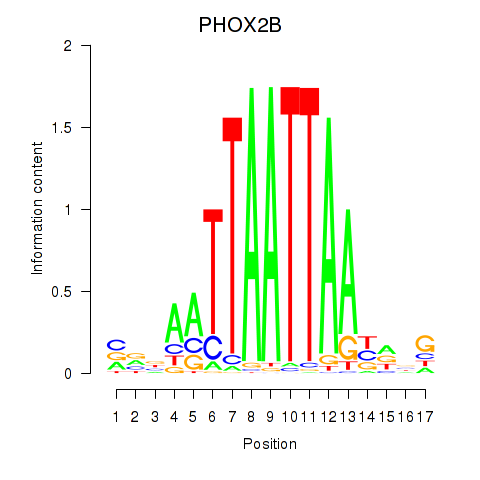
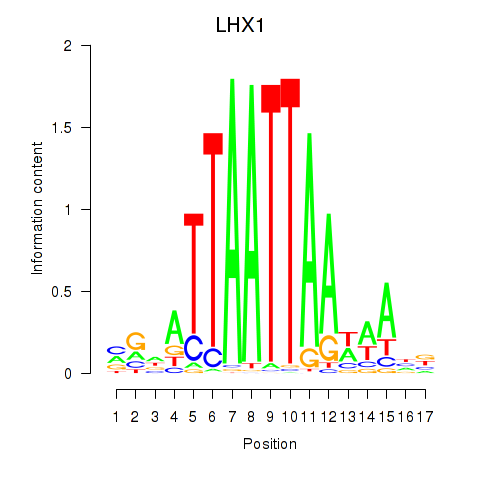
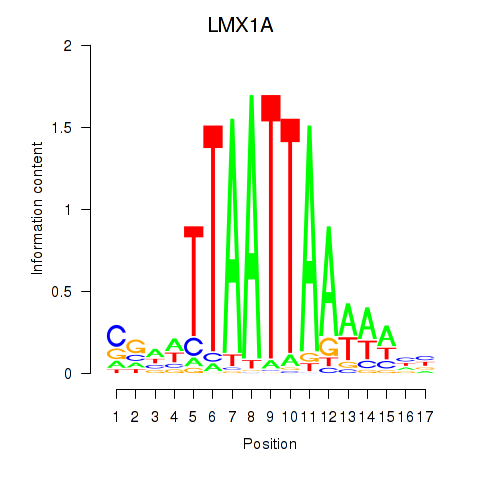
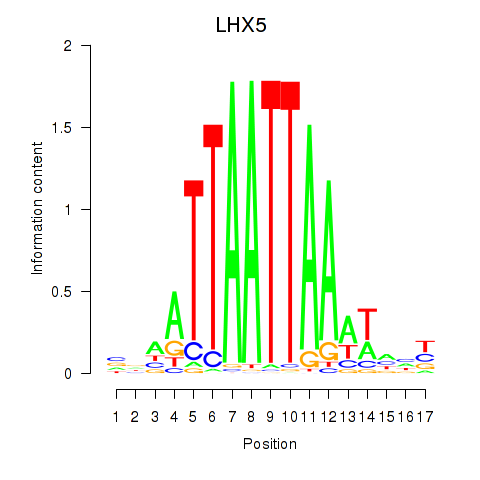
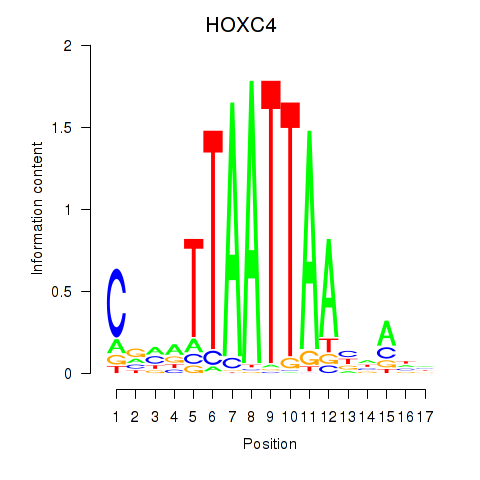
Transcription factors associated with OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTP
|
ENSG00000171540.8 | orthopedia homeobox |
|
PHOX2B
|
ENSG00000109132.7 | paired like homeobox 2B |
|
LHX1
|
ENSG00000273706.5 | LIM homeobox 1 |
|
LMX1A
|
ENSG00000162761.14 | LIM homeobox transcription factor 1 alpha |
|
LHX5
|
ENSG00000089116.4 | LIM homeobox 5 |
|
HOXC4
|
ENSG00000198353.8 | homeobox C4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OTP | hg38_v1_chr5_-_77638713_77638713 | -0.86 | 6.4e-03 | Click! |
| LMX1A | hg38_v1_chr1_-_165355746_165355779 | -0.66 | 7.4e-02 | Click! |
| HOXC4 | hg38_v1_chr12_+_54053815_54053875 | -0.47 | 2.4e-01 | Click! |
| LHX5 | hg38_v1_chr12_-_113471851_113471876 | -0.29 | 4.8e-01 | Click! |
| PHOX2B | hg38_v1_chr4_-_41748713_41748731 | -0.24 | 5.7e-01 | Click! |
Activity profile of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
Sorted Z-values of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_49513353 | 2.83 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr11_-_63608542 | 2.67 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr5_-_20575850 | 2.42 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr17_+_1771688 | 2.08 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr19_+_49513154 | 2.05 |
ENST00000426395.7
ENST00000600273.5 ENST00000599988.5 |
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr2_+_151357583 | 1.80 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr5_+_95731300 | 1.69 |
ENST00000379982.8
|
RHOBTB3
|
Rho related BTB domain containing 3 |
| chr6_+_113857333 | 1.48 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr20_-_35147285 | 1.41 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr3_+_45886501 | 1.28 |
ENST00000395963.2
|
CCR9
|
C-C motif chemokine receptor 9 |
| chr3_+_157436842 | 1.23 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr9_-_96778053 | 1.07 |
ENST00000375231.5
ENST00000223428.9 |
ZNF510
|
zinc finger protein 510 |
| chr16_-_66549839 | 1.01 |
ENST00000527800.6
ENST00000677555.1 ENST00000563369.6 |
TK2
|
thymidine kinase 2 |
| chrX_+_10158448 | 1.00 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr1_-_91906280 | 0.99 |
ENST00000370399.6
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr7_+_134843884 | 0.97 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr16_-_66550091 | 0.96 |
ENST00000564917.5
ENST00000677420.1 |
TK2
|
thymidine kinase 2 |
| chr3_-_165078480 | 0.94 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr16_-_66550005 | 0.90 |
ENST00000527284.6
|
TK2
|
thymidine kinase 2 |
| chr3_+_12287899 | 0.86 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr3_+_12287859 | 0.84 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr2_-_40453438 | 0.79 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr4_+_168497066 | 0.78 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr19_-_13953302 | 0.78 |
ENST00000585607.1
ENST00000538517.6 ENST00000587458.1 ENST00000538371.6 |
PODNL1
|
podocan like 1 |
| chr11_-_122116215 | 0.77 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr1_+_78303865 | 0.76 |
ENST00000370758.5
|
PTGFR
|
prostaglandin F receptor |
| chrX_+_22032301 | 0.76 |
ENST00000379374.5
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr14_-_50561119 | 0.73 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_-_55722857 | 0.73 |
ENST00000424486.3
|
TMEM100
|
transmembrane protein 100 |
| chr1_-_56579555 | 0.72 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr7_+_138460238 | 0.69 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr20_+_45416551 | 0.68 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr14_+_34993240 | 0.68 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr5_-_88823763 | 0.65 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr16_-_66550142 | 0.64 |
ENST00000417693.8
ENST00000299697.12 ENST00000451102.7 |
TK2
|
thymidine kinase 2 |
| chr12_+_8157034 | 0.62 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr5_+_141382702 | 0.60 |
ENST00000617050.1
ENST00000518325.2 |
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr13_-_37598750 | 0.57 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr4_+_168497044 | 0.56 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_-_28282086 | 0.54 |
ENST00000375719.7
ENST00000375732.5 |
MPP7
|
membrane palmitoylated protein 7 |
| chrX_+_43656289 | 0.54 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr8_-_42768602 | 0.51 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr3_-_142000353 | 0.51 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr12_+_26011713 | 0.50 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr20_-_18497218 | 0.49 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase |
| chr5_-_111758061 | 0.47 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr1_-_113871665 | 0.44 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr12_-_54259531 | 0.43 |
ENST00000550411.5
ENST00000439541.6 |
CBX5
|
chromobox 5 |
| chr19_-_43883964 | 0.43 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr17_+_7407838 | 0.41 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr3_-_9952337 | 0.41 |
ENST00000411976.2
ENST00000412055.6 |
PRRT3
|
proline rich transmembrane protein 3 |
| chr11_+_92969651 | 0.41 |
ENST00000257068.3
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr15_+_92900338 | 0.41 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chrX_+_51743395 | 0.40 |
ENST00000340438.6
|
GSPT2
|
G1 to S phase transition 2 |
| chr10_+_92691897 | 0.39 |
ENST00000492654.3
|
HHEX
|
hematopoietically expressed homeobox |
| chr3_-_157503574 | 0.39 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr20_+_15196834 | 0.38 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr9_+_72577788 | 0.38 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr9_+_122371014 | 0.38 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr15_-_19988117 | 0.38 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr4_+_70242583 | 0.37 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr9_+_72577369 | 0.37 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr5_-_126595237 | 0.37 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr6_+_136038195 | 0.36 |
ENST00000615259.4
|
PDE7B
|
phosphodiesterase 7B |
| chr17_+_41966814 | 0.35 |
ENST00000393888.1
ENST00000441615.2 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr4_+_112647059 | 0.34 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr10_-_27240505 | 0.33 |
ENST00000375888.5
ENST00000676732.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr3_+_138621225 | 0.33 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_+_64404338 | 0.33 |
ENST00000332707.10
|
XPOT
|
exportin for tRNA |
| chr3_+_69763549 | 0.32 |
ENST00000472437.5
|
MITF
|
melanocyte inducing transcription factor |
| chr13_-_109786567 | 0.32 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr5_-_127073467 | 0.31 |
ENST00000607731.5
ENST00000509733.7 ENST00000296662.10 ENST00000535381.6 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr1_+_177170916 | 0.31 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr5_+_154941063 | 0.30 |
ENST00000523037.6
ENST00000265229.12 ENST00000439747.7 ENST00000522038.5 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr21_+_32298849 | 0.30 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr8_+_103372388 | 0.30 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr1_+_158461574 | 0.29 |
ENST00000641432.1
ENST00000641460.1 ENST00000641535.1 ENST00000641971.1 |
OR10K1
|
olfactory receptor family 10 subfamily K member 1 |
| chr1_+_77918128 | 0.29 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr10_+_92691813 | 0.28 |
ENST00000472590.6
|
HHEX
|
hematopoietically expressed homeobox |
| chr20_+_58907981 | 0.27 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr17_-_445939 | 0.27 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr7_-_22822829 | 0.26 |
ENST00000358435.9
ENST00000621567.1 |
TOMM7
|
translocase of outer mitochondrial membrane 7 |
| chrX_+_85003863 | 0.26 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like |
| chr7_-_22822779 | 0.25 |
ENST00000372879.8
|
TOMM7
|
translocase of outer mitochondrial membrane 7 |
| chr9_-_26947222 | 0.24 |
ENST00000520884.5
ENST00000397292.8 |
PLAA
|
phospholipase A2 activating protein |
| chrX_-_101617921 | 0.24 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr1_+_201648136 | 0.24 |
ENST00000367296.8
|
NAV1
|
neuron navigator 1 |
| chr3_+_69763726 | 0.24 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr3_+_138621207 | 0.22 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_-_69860138 | 0.22 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1 |
| chr12_+_130953898 | 0.21 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr10_+_18260715 | 0.21 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr2_+_108621260 | 0.20 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr19_-_51417791 | 0.20 |
ENST00000353836.9
|
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr20_-_57690624 | 0.20 |
ENST00000414037.5
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr3_-_64225436 | 0.20 |
ENST00000638394.2
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr2_-_74391837 | 0.20 |
ENST00000417090.1
ENST00000409868.5 ENST00000680606.1 |
DCTN1
|
dynactin subunit 1 |
| chr8_+_23288081 | 0.19 |
ENST00000265806.12
ENST00000519952.6 ENST00000518840.6 |
R3HCC1
|
R3H domain and coiled-coil containing 1 |
| chr3_-_149576203 | 0.19 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr7_+_130293134 | 0.19 |
ENST00000445470.6
ENST00000492072.5 ENST00000222482.10 ENST00000473956.5 ENST00000493259.5 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr1_+_212791828 | 0.19 |
ENST00000532324.5
ENST00000530441.5 ENST00000526641.5 ENST00000531963.5 ENST00000366973.8 ENST00000366974.9 ENST00000526997.5 ENST00000488246.6 |
TATDN3
|
TatD DNase domain containing 3 |
| chr15_+_41286011 | 0.19 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr3_-_157503375 | 0.19 |
ENST00000362010.7
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr14_+_55027200 | 0.19 |
ENST00000395472.2
ENST00000555846.2 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr18_-_35290184 | 0.18 |
ENST00000589178.5
ENST00000592278.1 ENST00000333206.10 ENST00000592211.1 ENST00000420878.7 ENST00000610712.1 ENST00000586922.2 |
ZSCAN30
ENSG00000268573.1
|
zinc finger and SCAN domain containing 30 novel transcript |
| chr16_-_66550112 | 0.18 |
ENST00000544898.6
ENST00000620035.5 ENST00000545043.6 |
TK2
|
thymidine kinase 2 |
| chr9_-_111036207 | 0.18 |
ENST00000541779.5
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr17_-_8295342 | 0.17 |
ENST00000579192.5
ENST00000577745.2 |
SLC25A35
|
solute carrier family 25 member 35 |
| chr17_+_18183052 | 0.17 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr17_+_41966787 | 0.17 |
ENST00000393892.8
ENST00000587679.1 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr13_+_45702306 | 0.17 |
ENST00000533564.1
ENST00000310521.6 |
CBY2
|
chibby family member 2 |
| chr20_+_33283205 | 0.17 |
ENST00000253354.2
|
BPIFB1
|
BPI fold containing family B member 1 |
| chr14_+_24120956 | 0.16 |
ENST00000558325.2
|
ENSG00000259371.2
|
novel protein |
| chr19_-_14835252 | 0.16 |
ENST00000641666.1
ENST00000642030.1 ENST00000642000.1 |
OR7C1
|
olfactory receptor family 7 subfamily C member 1 |
| chr6_+_26402289 | 0.16 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr15_+_92900189 | 0.16 |
ENST00000626874.2
ENST00000627622.1 ENST00000629346.2 ENST00000628375.2 ENST00000420239.7 ENST00000394196.9 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr14_+_55027576 | 0.16 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr6_+_47781982 | 0.16 |
ENST00000489301.6
ENST00000638973.1 ENST00000371211.6 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr3_-_157503339 | 0.16 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr13_+_45702411 | 0.15 |
ENST00000610924.1
|
CBY2
|
chibby family member 2 |
| chr19_+_52297157 | 0.15 |
ENST00000595962.6
ENST00000598016.5 ENST00000334564.11 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr17_+_76467597 | 0.15 |
ENST00000392492.8
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr6_+_26440472 | 0.14 |
ENST00000494393.5
ENST00000482451.5 ENST00000471353.5 ENST00000361232.7 ENST00000487627.5 ENST00000496719.1 ENST00000244519.7 ENST00000490254.5 ENST00000487272.1 |
BTN3A3
|
butyrophilin subfamily 3 member A3 |
| chr19_-_14835162 | 0.14 |
ENST00000322301.5
|
OR7A5
|
olfactory receptor family 7 subfamily A member 5 |
| chr7_-_143902114 | 0.14 |
ENST00000355951.2
ENST00000479870.6 ENST00000478172.1 |
TCAF1
|
TRPM8 channel associated factor 1 |
| chr13_+_53028806 | 0.14 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chrX_+_55717733 | 0.13 |
ENST00000414239.5
ENST00000374941.9 |
RRAGB
|
Ras related GTP binding B |
| chr7_+_8433602 | 0.13 |
ENST00000405863.6
|
NXPH1
|
neurexophilin 1 |
| chr11_-_117316230 | 0.13 |
ENST00000313005.11
ENST00000528053.5 |
BACE1
|
beta-secretase 1 |
| chr2_+_102473219 | 0.13 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr6_-_41071825 | 0.13 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr11_-_18046262 | 0.13 |
ENST00000682019.1
|
TPH1
|
tryptophan hydroxylase 1 |
| chr12_+_26195647 | 0.13 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr13_-_61415508 | 0.13 |
ENST00000409204.4
|
PCDH20
|
protocadherin 20 |
| chrX_+_55717796 | 0.12 |
ENST00000262850.7
|
RRAGB
|
Ras related GTP binding B |
| chr2_-_20823048 | 0.12 |
ENST00000402479.6
ENST00000237822.8 ENST00000626491.2 ENST00000432947.1 ENST00000403006.6 ENST00000419825.2 ENST00000381090.7 ENST00000412261.5 ENST00000619656.4 ENST00000541941.5 ENST00000440866.6 ENST00000435420.6 |
LDAH
|
lipid droplet associated hydrolase |
| chr1_-_109075944 | 0.12 |
ENST00000338366.6
|
TAF13
|
TATA-box binding protein associated factor 13 |
| chr17_+_68515399 | 0.12 |
ENST00000588188.6
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr1_+_117001744 | 0.12 |
ENST00000256652.8
ENST00000682167.1 ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr12_+_8509460 | 0.12 |
ENST00000382064.6
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chrX_+_109535775 | 0.12 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr7_+_150323239 | 0.12 |
ENST00000323078.7
ENST00000493307.1 ENST00000359623.9 |
LRRC61
|
leucine rich repeat containing 61 |
| chr19_+_21142024 | 0.12 |
ENST00000600692.5
ENST00000599296.5 ENST00000594425.5 ENST00000311048.11 |
ZNF431
|
zinc finger protein 431 |
| chr1_+_248508073 | 0.11 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr2_-_74392025 | 0.11 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr9_+_93234923 | 0.11 |
ENST00000411624.5
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr15_+_64387828 | 0.11 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr18_+_34976928 | 0.10 |
ENST00000591734.5
ENST00000413393.5 ENST00000589180.5 ENST00000587359.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr20_-_31390483 | 0.10 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chr14_-_106470788 | 0.10 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr8_-_89984231 | 0.10 |
ENST00000517337.1
ENST00000409330.5 |
NBN
|
nibrin |
| chr11_-_102705737 | 0.10 |
ENST00000260229.5
|
MMP27
|
matrix metallopeptidase 27 |
| chr15_+_21579912 | 0.10 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr8_-_115492221 | 0.10 |
ENST00000518018.1
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr16_+_28553908 | 0.10 |
ENST00000317058.8
|
SGF29
|
SAGA complex associated factor 29 |
| chr6_-_111793871 | 0.09 |
ENST00000368667.6
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr3_-_99850976 | 0.09 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like |
| chr14_-_106658251 | 0.09 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr3_-_157499538 | 0.09 |
ENST00000392832.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr9_-_28670285 | 0.09 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr6_+_122471913 | 0.09 |
ENST00000615438.4
ENST00000392491.6 |
PKIB
|
cAMP-dependent protein kinase inhibitor beta |
| chr14_+_21997531 | 0.09 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr12_+_8513499 | 0.09 |
ENST00000299665.3
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chr3_+_141387801 | 0.08 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_-_191282383 | 0.08 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr2_-_190250503 | 0.08 |
ENST00000409820.2
ENST00000410045.5 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr7_+_134127299 | 0.08 |
ENST00000645682.1
ENST00000285928.2 |
LRGUK
|
leucine rich repeats and guanylate kinase domain containing |
| chr16_-_71809049 | 0.08 |
ENST00000569748.5
ENST00000570017.1 ENST00000393512.7 |
AP1G1
|
adaptor related protein complex 1 subunit gamma 1 |
| chr1_-_212791762 | 0.08 |
ENST00000626725.1
ENST00000366977.8 ENST00000366976.3 |
NSL1
|
NSL1 component of MIS12 kinetochore complex |
| chr7_-_122098831 | 0.08 |
ENST00000681213.1
ENST00000679419.1 |
AASS
|
aminoadipate-semialdehyde synthase |
| chr3_-_146528750 | 0.08 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_+_44799187 | 0.07 |
ENST00000425755.5
|
KIF15
|
kinesin family member 15 |
| chr3_-_186544377 | 0.07 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S |
| chr4_+_186069144 | 0.07 |
ENST00000513189.1
ENST00000296795.8 |
TLR3
|
toll like receptor 3 |
| chr20_-_35529618 | 0.07 |
ENST00000246199.5
ENST00000424444.1 ENST00000374345.8 ENST00000444723.3 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr3_+_140678041 | 0.07 |
ENST00000286349.4
|
TRIM42
|
tripartite motif containing 42 |
| chr1_-_158426237 | 0.07 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr1_-_100894818 | 0.07 |
ENST00000370114.8
|
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr1_-_100894775 | 0.07 |
ENST00000416479.1
ENST00000370113.7 |
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr9_+_34652167 | 0.07 |
ENST00000441545.7
ENST00000553620.5 |
IL11RA
|
interleukin 11 receptor subunit alpha |
| chr12_+_9971402 | 0.07 |
ENST00000304361.9
ENST00000396507.7 ENST00000434319.6 |
CLEC12A
|
C-type lectin domain family 12 member A |
| chr4_+_26343156 | 0.07 |
ENST00000680928.1
ENST00000681260.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_+_70226116 | 0.07 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr3_+_141738263 | 0.07 |
ENST00000480908.1
ENST00000393000.3 ENST00000273480.4 |
RNF7
|
ring finger protein 7 |
| chr1_-_157700738 | 0.07 |
ENST00000368186.9
ENST00000496769.1 ENST00000368184.8 |
FCRL3
|
Fc receptor like 3 |
| chr4_-_151227881 | 0.07 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr2_+_30447211 | 0.06 |
ENST00000466477.5
ENST00000465200.5 ENST00000319406.8 ENST00000379509.8 ENST00000488144.5 ENST00000465538.5 ENST00000309052.8 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr5_-_131165231 | 0.06 |
ENST00000675100.1
ENST00000304043.10 ENST00000513012.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr5_-_131165272 | 0.06 |
ENST00000675491.1
ENST00000506908.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr5_-_177462379 | 0.06 |
ENST00000512501.1
|
DBN1
|
drebrin 1 |
| chr9_-_74952904 | 0.06 |
ENST00000376854.6
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr9_+_108862255 | 0.06 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A |
| chr18_-_36798482 | 0.06 |
ENST00000590258.2
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr8_-_92966129 | 0.06 |
ENST00000522925.5
ENST00000522903.5 ENST00000537541.1 ENST00000521988.6 ENST00000518748.5 ENST00000519069.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_-_118359105 | 0.06 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr6_-_169250825 | 0.06 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr7_-_87475839 | 0.06 |
ENST00000359206.8
|
ABCB4
|
ATP binding cassette subfamily B member 4 |
| chr15_-_75455767 | 0.05 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chrX_+_77910656 | 0.05 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr19_-_44401575 | 0.05 |
ENST00000591679.5
ENST00000544719.6 ENST00000614994.5 ENST00000585868.1 ENST00000588212.1 |
ZNF285
ENSG00000267173.1
|
zinc finger protein 285 novel protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.6 | 3.7 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.5 | 1.4 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.4 | 1.3 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.4 | 1.2 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.3 | 2.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.2 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) |
| 0.2 | 1.7 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.7 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.2 | 0.8 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 0.7 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.6 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.7 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.4 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.3 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 0.4 | GO:1902523 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.4 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.8 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.8 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 1.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.7 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 1.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0042435 | serotonin biosynthetic process(GO:0042427) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.1 | GO:0072573 | tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0031860 | regulation of DNA-dependent DNA replication initiation(GO:0030174) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 1.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:1905237 | response to cyclosporin A(GO:1905237) positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) regulation of dendritic cell cytokine production(GO:0002730) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.8 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 2.3 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 2.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 3.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 3.7 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.3 | 0.8 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 0.9 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 1.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.0 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.8 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |