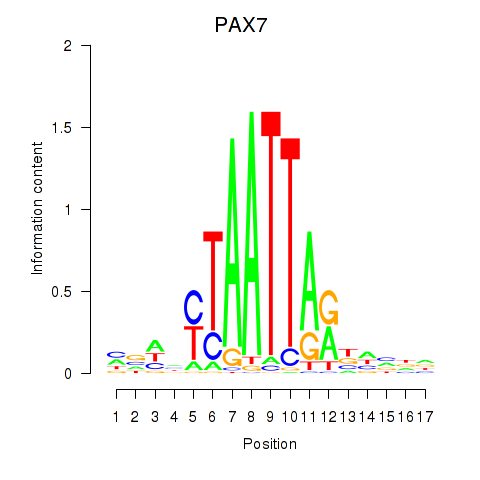
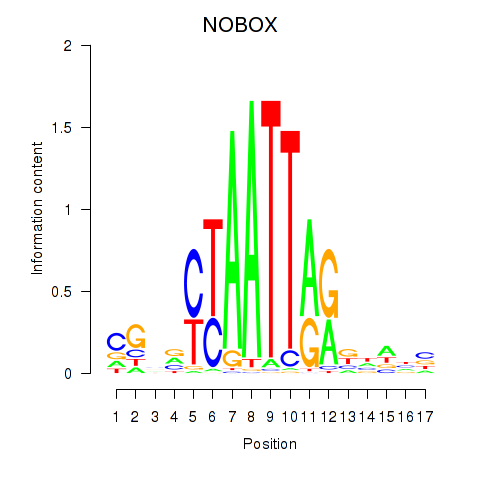
|
chr12_+_26195313
Show fit
|
0.66 |
ENST00000422622.3
|
SSPN
|
sarcospan
|
|
chr3_-_165837412
Show fit
|
0.60 |
ENST00000479451.5
ENST00000488954.1
ENST00000264381.8
|
BCHE
|
butyrylcholinesterase
|
|
chr17_+_41237998
Show fit
|
0.44 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8
|
|
chr8_+_104223320
Show fit
|
0.42 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr4_+_94974984
Show fit
|
0.38 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr9_-_92404559
Show fit
|
0.38 |
ENST00000262551.8
ENST00000375561.10
|
OGN
|
osteoglycin
|
|
chr5_+_55851378
Show fit
|
0.36 |
ENST00000396836.6
ENST00000359040.10
|
IL31RA
|
interleukin 31 receptor A
|
|
chr12_+_26195543
Show fit
|
0.35 |
ENST00000242729.7
|
SSPN
|
sarcospan
|
|
chr13_-_85799400
Show fit
|
0.34 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6
|
|
chr4_-_137532452
Show fit
|
0.33 |
ENST00000412923.6
ENST00000511115.5
ENST00000344876.9
ENST00000507846.5
ENST00000510305.5
ENST00000611581.1
|
PCDH18
|
protocadherin 18
|
|
chr6_+_47656436
Show fit
|
0.32 |
ENST00000507065.5
ENST00000296862.5
|
ADGRF2
|
adhesion G protein-coupled receptor F2
|
|
chr18_-_3219849
Show fit
|
0.29 |
ENST00000261606.11
|
MYOM1
|
myomesin 1
|
|
chr8_+_104223344
Show fit
|
0.28 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr3_-_151316795
Show fit
|
0.28 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87
|
|
chr15_+_94355956
Show fit
|
0.27 |
ENST00000557742.1
|
MCTP2
|
multiple C2 and transmembrane domain containing 2
|
|
chr10_-_104085847
Show fit
|
0.25 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain
|
|
chr1_-_161021096
Show fit
|
0.24 |
ENST00000537746.1
ENST00000368026.11
|
F11R
|
F11 receptor
|
|
chr2_+_68734773
Show fit
|
0.23 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr19_+_35106510
Show fit
|
0.23 |
ENST00000648240.1
|
ENSG00000285526.1
|
novel protein
|
|
chr18_+_63587297
Show fit
|
0.22 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13
|
|
chr1_+_81306096
Show fit
|
0.22 |
ENST00000370721.5
ENST00000370727.5
ENST00000370725.5
ENST00000370723.5
ENST00000370728.5
ENST00000370730.5
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr4_+_85778681
Show fit
|
0.22 |
ENST00000395183.6
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr3_+_156291834
Show fit
|
0.22 |
ENST00000389634.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1
|
|
chr18_+_63587336
Show fit
|
0.22 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13
|
|
chr7_-_14903319
Show fit
|
0.22 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta
|
|
chrX_+_136205982
Show fit
|
0.22 |
ENST00000628568.1
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_26402237
Show fit
|
0.21 |
ENST00000476549.6
ENST00000450085.6
ENST00000425234.6
ENST00000427334.5
ENST00000506698.1
ENST00000289361.11
|
BTN3A1
|
butyrophilin subfamily 3 member A1
|
|
chr17_+_41226648
Show fit
|
0.21 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr9_+_78297143
Show fit
|
0.21 |
ENST00000347159.6
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr1_+_186828941
Show fit
|
0.21 |
ENST00000367466.4
|
PLA2G4A
|
phospholipase A2 group IVA
|
|
chr14_-_74493322
Show fit
|
0.20 |
ENST00000553490.5
ENST00000557510.5
|
NPC2
|
NPC intracellular cholesterol transporter 2
|
|
chrX_+_43656289
Show fit
|
0.20 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A
|
|
chr18_+_46946821
Show fit
|
0.20 |
ENST00000245121.9
ENST00000356157.12
|
KATNAL2
|
katanin catalytic subunit A1 like 2
|
|
chr1_+_27934980
Show fit
|
0.19 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B
|
|
chr14_-_74493275
Show fit
|
0.19 |
ENST00000541064.5
|
NPC2
|
NPC intracellular cholesterol transporter 2
|
|
chr14_-_74493291
Show fit
|
0.19 |
ENST00000238633.6
ENST00000555619.6
ENST00000434013.6
|
NPC2
|
NPC intracellular cholesterol transporter 2
|
|
chr6_-_155455830
Show fit
|
0.18 |
ENST00000159060.3
|
NOX3
|
NADPH oxidase 3
|
|
chr12_-_9999176
Show fit
|
0.17 |
ENST00000298527.10
ENST00000348658.4
|
CLEC1B
|
C-type lectin domain family 1 member B
|
|
chr22_-_21735744
Show fit
|
0.17 |
ENST00000403503.1
|
YPEL1
|
yippee like 1
|
|
chr1_+_110873135
Show fit
|
0.17 |
ENST00000271324.6
|
CD53
|
CD53 molecule
|
|
chr11_+_118077009
Show fit
|
0.17 |
ENST00000616579.4
ENST00000534111.5
|
TMPRSS4
|
transmembrane serine protease 4
|
|
chr10_+_46375619
Show fit
|
0.17 |
ENST00000584982.7
ENST00000613703.4
|
ANXA8L1
|
annexin A8 like 1
|
|
chr9_+_78297117
Show fit
|
0.16 |
ENST00000376588.4
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr5_-_83673544
Show fit
|
0.16 |
ENST00000503117.1
ENST00000510978.5
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr17_+_1771688
Show fit
|
0.16 |
ENST00000572048.1
ENST00000573763.1
|
SERPINF1
|
serpin family F member 1
|
|
chr10_+_46375645
Show fit
|
0.15 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1
|
|
chr4_+_112860912
Show fit
|
0.15 |
ENST00000671951.1
|
ANK2
|
ankyrin 2
|
|
chr14_+_22202561
Show fit
|
0.15 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2
|
|
chr4_+_112861053
Show fit
|
0.15 |
ENST00000672221.1
|
ANK2
|
ankyrin 2
|
|
chr4_+_112860981
Show fit
|
0.15 |
ENST00000671704.1
|
ANK2
|
ankyrin 2
|
|
chr16_-_28623560
Show fit
|
0.15 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1
|
|
chr2_-_96034916
Show fit
|
0.15 |
ENST00000359548.8
ENST00000439254.1
ENST00000453542.5
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial
|
|
chr17_-_7263959
Show fit
|
0.14 |
ENST00000571932.2
|
CLDN7
|
claudin 7
|
|
chr11_+_118077067
Show fit
|
0.14 |
ENST00000522307.5
ENST00000523251.5
ENST00000437212.8
ENST00000522824.5
ENST00000522151.5
|
TMPRSS4
|
transmembrane serine protease 4
|
|
chr4_+_2418932
Show fit
|
0.14 |
ENST00000635017.1
|
CFAP99
|
cilia and flagella associated protein 99
|
|
chr12_+_107318395
Show fit
|
0.14 |
ENST00000420571.6
ENST00000280758.10
|
BTBD11
|
BTB domain containing 11
|
|
chr1_+_160400543
Show fit
|
0.14 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2
|
|
chr6_+_26124161
Show fit
|
0.14 |
ENST00000377791.4
ENST00000602637.1
|
H2AC6
|
H2A clustered histone 6
|
|
chr5_+_67004618
Show fit
|
0.14 |
ENST00000261569.11
ENST00000436277.5
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr18_-_55423757
Show fit
|
0.13 |
ENST00000675707.1
|
TCF4
|
transcription factor 4
|
|
chr1_-_183590876
Show fit
|
0.13 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr15_+_73926443
Show fit
|
0.13 |
ENST00000261921.8
|
LOXL1
|
lysyl oxidase like 1
|
|
chrX_+_139530730
Show fit
|
0.12 |
ENST00000218099.7
|
F9
|
coagulation factor IX
|
|
chr20_+_15196834
Show fit
|
0.12 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2
|
|
chr2_+_165239432
Show fit
|
0.12 |
ENST00000636071.2
ENST00000636985.2
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2
|
|
chr10_-_130110806
Show fit
|
0.12 |
ENST00000637128.2
|
C10orf143
|
chromosome 10 open reading frame 143
|
|
chr6_+_26440472
Show fit
|
0.11 |
ENST00000494393.5
ENST00000482451.5
ENST00000471353.5
ENST00000361232.7
ENST00000487627.5
ENST00000496719.1
ENST00000244519.7
ENST00000490254.5
ENST00000487272.1
|
BTN3A3
|
butyrophilin subfamily 3 member A3
|
|
chr6_+_26365215
Show fit
|
0.11 |
ENST00000527422.5
ENST00000356386.6
ENST00000396948.5
|
BTN3A2
|
butyrophilin subfamily 3 member A2
|
|
chr2_+_188292814
Show fit
|
0.11 |
ENST00000409580.5
ENST00000409637.7
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1
|
|
chr15_+_58138368
Show fit
|
0.11 |
ENST00000219919.9
ENST00000536493.1
|
AQP9
|
aquaporin 9
|
|
chr9_-_92536031
Show fit
|
0.11 |
ENST00000344604.9
ENST00000375540.5
|
ECM2
|
extracellular matrix protein 2
|
|
chr11_-_120120880
Show fit
|
0.10 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29
|
|
chr6_+_26365176
Show fit
|
0.10 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2
|
|
chr15_+_76336755
Show fit
|
0.10 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2
|
|
chr2_+_165239388
Show fit
|
0.10 |
ENST00000424833.5
ENST00000375437.7
ENST00000631182.3
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2
|
|
chr19_+_11374658
Show fit
|
0.10 |
ENST00000674460.1
ENST00000312423.4
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1
|
|
chr10_+_89392546
Show fit
|
0.10 |
ENST00000546318.2
ENST00000371804.4
|
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1
|
|
chr3_-_142029108
Show fit
|
0.10 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr11_+_57597563
Show fit
|
0.09 |
ENST00000619430.2
ENST00000457869.1
ENST00000340687.10
ENST00000278407.9
ENST00000378323.8
ENST00000378324.6
ENST00000403558.1
|
SERPING1
|
serpin family G member 1
|
|
chr2_+_68734861
Show fit
|
0.09 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr5_+_140841183
Show fit
|
0.09 |
ENST00000378123.4
ENST00000531613.2
|
PCDHA8
|
protocadherin alpha 8
|
|
chr16_+_11965193
Show fit
|
0.09 |
ENST00000053243.6
ENST00000396495.3
|
TNFRSF17
|
TNF receptor superfamily member 17
|
|
chr12_+_26195647
Show fit
|
0.09 |
ENST00000535504.1
|
SSPN
|
sarcospan
|
|
chr9_+_72577939
Show fit
|
0.09 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1
|
|
chr11_-_59615673
Show fit
|
0.09 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein
|
|
chr3_-_105868964
Show fit
|
0.08 |
ENST00000394030.8
|
CBLB
|
Cbl proto-oncogene B
|
|
chr17_-_42185452
Show fit
|
0.08 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor
|
|
chr4_-_25863537
Show fit
|
0.08 |
ENST00000502949.5
ENST00000264868.9
ENST00000513691.1
ENST00000514872.1
|
SEL1L3
|
SEL1L family member 3
|
|
chrX_+_131083706
Show fit
|
0.08 |
ENST00000370921.1
|
ARHGAP36
|
Rho GTPase activating protein 36
|
|
chr11_+_95789965
Show fit
|
0.08 |
ENST00000537677.5
|
CEP57
|
centrosomal protein 57
|
|
chr1_+_151766655
Show fit
|
0.08 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr1_-_12848720
Show fit
|
0.08 |
ENST00000317869.7
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C like 1
|
|
chr14_+_103715724
Show fit
|
0.08 |
ENST00000216602.10
|
ZFYVE21
|
zinc finger FYVE-type containing 21
|
|
chr18_+_58341038
Show fit
|
0.08 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr9_-_4299547
Show fit
|
0.08 |
ENST00000682749.1
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_+_186694007
Show fit
|
0.08 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B
|
|
chr20_-_51802509
Show fit
|
0.08 |
ENST00000371539.7
ENST00000217086.9
|
SALL4
|
spalt like transcription factor 4
|
|
chr20_-_51802433
Show fit
|
0.08 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4
|
|
chrX_-_15314543
Show fit
|
0.08 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr14_+_103715767
Show fit
|
0.08 |
ENST00000311141.7
|
ZFYVE21
|
zinc finger FYVE-type containing 21
|
|
chrX_-_17861236
Show fit
|
0.07 |
ENST00000331511.5
ENST00000415486.7
ENST00000451717.6
ENST00000545871.1
|
RAI2
|
retinoic acid induced 2
|
|
chr1_-_16978276
Show fit
|
0.07 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2
|
|
chr19_-_3557563
Show fit
|
0.07 |
ENST00000389395.7
ENST00000355415.7
|
MFSD12
|
major facilitator superfamily domain containing 12
|
|
chr8_-_30812867
Show fit
|
0.07 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta
|
|
chr1_+_234373667
Show fit
|
0.07 |
ENST00000366613.1
ENST00000366612.1
|
COA6
|
cytochrome c oxidase assembly factor 6
|
|
chr16_-_3443446
Show fit
|
0.07 |
ENST00000301744.7
|
ZNF597
|
zinc finger protein 597
|
|
chr4_-_89029881
Show fit
|
0.07 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A
|
|
chr10_-_97445850
Show fit
|
0.07 |
ENST00000477692.6
ENST00000485122.6
ENST00000370886.9
ENST00000370885.8
ENST00000370902.8
ENST00000370884.5
|
EXOSC1
|
exosome component 1
|
|
chr1_+_207053229
Show fit
|
0.07 |
ENST00000367080.8
ENST00000367079.3
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr10_-_97687191
Show fit
|
0.07 |
ENST00000370626.4
|
AVPI1
|
arginine vasopressin induced 1
|
|
chr3_-_105869035
Show fit
|
0.07 |
ENST00000447441.6
ENST00000403724.5
ENST00000405772.5
|
CBLB
|
Cbl proto-oncogene B
|
|
chr2_-_165203870
Show fit
|
0.07 |
ENST00000639244.1
ENST00000409101.7
ENST00000668657.1
|
SCN3A
|
sodium voltage-gated channel alpha subunit 3
|
|
chr5_+_40841308
Show fit
|
0.07 |
ENST00000381677.4
ENST00000254691.10
|
CARD6
|
caspase recruitment domain family member 6
|
|
chr17_-_42676980
Show fit
|
0.07 |
ENST00000587627.1
ENST00000591022.6
ENST00000293349.10
|
PLEKHH3
|
pleckstrin homology, MyTH4 and FERM domain containing H3
|
|
chr4_-_99435134
Show fit
|
0.07 |
ENST00000476959.5
ENST00000482593.5
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr7_+_93906557
Show fit
|
0.06 |
ENST00000248572.10
ENST00000429473.1
ENST00000430875.1
ENST00000428834.1
|
GNGT1
|
G protein subunit gamma transducin 1
|
|
chr6_-_27132750
Show fit
|
0.06 |
ENST00000607124.1
ENST00000339812.3
|
H2BC11
|
H2B clustered histone 11
|
|
chr12_+_66302486
Show fit
|
0.06 |
ENST00000247815.9
|
HELB
|
DNA helicase B
|
|
chr4_+_186069144
Show fit
|
0.06 |
ENST00000513189.1
ENST00000296795.8
|
TLR3
|
toll like receptor 3
|
|
chr12_-_114403898
Show fit
|
0.06 |
ENST00000526441.1
|
TBX5
|
T-box transcription factor 5
|
|
chr2_+_90209873
Show fit
|
0.06 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43
|
|
chr4_+_154781213
Show fit
|
0.06 |
ENST00000514866.5
ENST00000281722.8
|
RBM46
|
RNA binding motif protein 46
|
|
chr19_+_55769118
Show fit
|
0.06 |
ENST00000341750.5
|
RFPL4AL1
|
ret finger protein like 4A like 1
|
|
chr8_-_108248700
Show fit
|
0.06 |
ENST00000220849.10
ENST00000678937.1
ENST00000678901.1
ENST00000678243.1
ENST00000677674.1
ENST00000521297.2
ENST00000677272.1
ENST00000522352.6
ENST00000678042.1
ENST00000519627.2
ENST00000521440.6
ENST00000678773.1
ENST00000676698.1
ENST00000518345.2
ENST00000677447.1
ENST00000676548.1
ENST00000519030.6
ENST00000518442.5
|
EIF3E
|
eukaryotic translation initiation factor 3 subunit E
|
|
chr4_+_87975667
Show fit
|
0.06 |
ENST00000237623.11
ENST00000682655.1
ENST00000508233.6
ENST00000360804.4
ENST00000395080.8
|
SPP1
|
secreted phosphoprotein 1
|
|
chr8_-_90082871
Show fit
|
0.06 |
ENST00000265431.7
|
CALB1
|
calbindin 1
|
|
chr3_-_171810432
Show fit
|
0.06 |
ENST00000356327.9
ENST00000351298.9
|
PLD1
|
phospholipase D1
|
|
chr9_-_92878018
Show fit
|
0.06 |
ENST00000332591.6
ENST00000375495.8
ENST00000395505.6
ENST00000395506.7
|
ZNF484
|
zinc finger protein 484
|
|
chr16_+_9091593
Show fit
|
0.06 |
ENST00000327827.12
|
C16orf72
|
chromosome 16 open reading frame 72
|
|
chr7_+_107583919
Show fit
|
0.06 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29
|
|
chrX_+_105948429
Show fit
|
0.06 |
ENST00000540278.1
|
NRK
|
Nik related kinase
|
|
chr4_-_73620391
Show fit
|
0.06 |
ENST00000395777.6
ENST00000307439.10
|
RASSF6
|
Ras association domain family member 6
|
|
chr7_+_71132123
Show fit
|
0.06 |
ENST00000333538.10
|
GALNT17
|
polypeptide N-acetylgalactosaminyltransferase 17
|
|
chr14_+_74084947
Show fit
|
0.06 |
ENST00000674221.1
ENST00000554938.2
|
LIN52
|
lin-52 DREAM MuvB core complex component
|
|
chrX_-_24672654
Show fit
|
0.06 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr4_+_76435216
Show fit
|
0.06 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3
|
|
chrX_+_22136552
Show fit
|
0.05 |
ENST00000682888.1
ENST00000684356.1
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked
|
|
chr6_+_132538290
Show fit
|
0.05 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9
|
|
chr18_-_63661884
Show fit
|
0.05 |
ENST00000332821.8
ENST00000283752.10
|
SERPINB3
|
serpin family B member 3
|
|
chr2_-_27663817
Show fit
|
0.05 |
ENST00000404798.6
|
SUPT7L
|
SPT7 like, STAGA complex subunit gamma
|
|
chr4_-_46124046
Show fit
|
0.05 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1
|
|
chr8_+_7881387
Show fit
|
0.05 |
ENST00000314357.4
|
DEFB103A
|
defensin beta 103A
|
|
chr1_+_100351698
Show fit
|
0.05 |
ENST00000644676.1
|
CDC14A
|
cell division cycle 14A
|
|
chr9_+_34652167
Show fit
|
0.05 |
ENST00000441545.7
ENST00000553620.5
|
IL11RA
|
interleukin 11 receptor subunit alpha
|
|
chr14_+_74493743
Show fit
|
0.05 |
ENST00000298818.12
ENST00000554924.1
ENST00000556816.6
|
ISCA2
|
iron-sulfur cluster assembly 2
|
|
chrX_+_71578435
Show fit
|
0.05 |
ENST00000373696.8
|
GCNA
|
germ cell nuclear acidic peptidase
|
|
chr7_-_14841267
Show fit
|
0.05 |
ENST00000406247.7
ENST00000399322.7
|
DGKB
|
diacylglycerol kinase beta
|
|
chr7_-_141014939
Show fit
|
0.05 |
ENST00000324787.10
ENST00000467334.1
|
MRPS33
|
mitochondrial ribosomal protein S33
|
|
chr5_+_142770367
Show fit
|
0.05 |
ENST00000645722.2
ENST00000274498.9
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr3_+_138621207
Show fit
|
0.05 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule
|
|
chr9_+_121567057
Show fit
|
0.05 |
ENST00000394340.7
ENST00000436835.5
ENST00000259371.6
|
DAB2IP
|
DAB2 interacting protein
|
|
chr4_-_46909206
Show fit
|
0.05 |
ENST00000396533.5
|
COX7B2
|
cytochrome c oxidase subunit 7B2
|
|
chr3_-_61251376
Show fit
|
0.05 |
ENST00000476844.5
ENST00000488467.5
ENST00000492590.6
ENST00000468189.5
|
FHIT
|
fragile histidine triad diadenosine triphosphatase
|
|
chr2_+_181985846
Show fit
|
0.05 |
ENST00000682840.1
ENST00000409137.7
ENST00000280295.7
|
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C
|
|
chr1_+_153967864
Show fit
|
0.05 |
ENST00000449724.5
ENST00000368607.8
ENST00000271889.8
|
CREB3L4
|
cAMP responsive element binding protein 3 like 4
|
|
chr7_+_100539188
Show fit
|
0.05 |
ENST00000300176.9
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr7_-_80919017
Show fit
|
0.05 |
ENST00000265361.8
|
SEMA3C
|
semaphorin 3C
|
|
chr6_-_26123910
Show fit
|
0.05 |
ENST00000314332.5
ENST00000396984.1
|
H2BC4
|
H2B clustered histone 4
|
|
chr7_-_57132425
Show fit
|
0.05 |
ENST00000319636.10
|
ZNF479
|
zinc finger protein 479
|
|
chr17_-_40665121
Show fit
|
0.05 |
ENST00000394052.5
|
KRT222
|
keratin 222
|
|
chr13_-_75366973
Show fit
|
0.05 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4
|
|
chr9_-_95317671
Show fit
|
0.05 |
ENST00000490972.7
ENST00000647778.1
ENST00000649611.1
ENST00000289081.8
|
FANCC
|
FA complementation group C
|
|
chr9_-_127771335
Show fit
|
0.05 |
ENST00000373276.7
ENST00000373277.8
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr8_-_134510182
Show fit
|
0.05 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing
|
|
chr19_-_10010492
Show fit
|
0.05 |
ENST00000264828.4
|
COL5A3
|
collagen type V alpha 3 chain
|
|
chr7_+_37920602
Show fit
|
0.05 |
ENST00000199448.9
ENST00000423717.1
|
EPDR1
|
ependymin related 1
|
|
chr3_-_112974912
Show fit
|
0.05 |
ENST00000440122.6
ENST00000490004.1
|
CD200R1
|
CD200 receptor 1
|
|
chr7_+_55887446
Show fit
|
0.04 |
ENST00000429591.4
|
ZNF713
|
zinc finger protein 713
|
|
chr16_+_3283443
Show fit
|
0.04 |
ENST00000572748.1
ENST00000219069.6
ENST00000573578.1
ENST00000574253.1
|
ZNF263
|
zinc finger protein 263
|
|
chr10_+_47322450
Show fit
|
0.04 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2
|
|
chr2_-_27663594
Show fit
|
0.04 |
ENST00000337768.10
ENST00000405491.5
ENST00000464789.2
ENST00000406540.5
|
SUPT7L
|
SPT7 like, STAGA complex subunit gamma
|
|
chr3_-_120285215
Show fit
|
0.04 |
ENST00000464295.6
|
GPR156
|
G protein-coupled receptor 156
|
|
chr8_+_12108172
Show fit
|
0.04 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D
|
|
chr6_-_111606260
Show fit
|
0.04 |
ENST00000340026.10
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_-_157820060
Show fit
|
0.04 |
ENST00000491942.1
ENST00000358292.7
|
FCRL1
|
Fc receptor like 1
|
|
chr11_+_118527463
Show fit
|
0.04 |
ENST00000302783.10
|
TTC36
|
tetratricopeptide repeat domain 36
|
|
chr8_-_85378105
Show fit
|
0.04 |
ENST00000521846.5
ENST00000523022.6
ENST00000524324.5
ENST00000519991.5
ENST00000520663.5
ENST00000517590.5
ENST00000522579.5
ENST00000522814.5
ENST00000522662.5
ENST00000523858.5
ENST00000519129.5
|
CA1
|
carbonic anhydrase 1
|
|
chr15_+_62561361
Show fit
|
0.04 |
ENST00000561311.5
|
TLN2
|
talin 2
|
|
chr19_+_7830188
Show fit
|
0.04 |
ENST00000270530.8
|
EVI5L
|
ecotropic viral integration site 5 like
|
|
chr14_+_56117702
Show fit
|
0.04 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2
|
|
chr15_-_26629083
Show fit
|
0.04 |
ENST00000400188.7
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3
|
|
chr10_+_97446137
Show fit
|
0.04 |
ENST00000370854.7
ENST00000393760.6
ENST00000414567.5
|
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16
|
|
chr11_-_117877463
Show fit
|
0.04 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr9_-_21368962
Show fit
|
0.04 |
ENST00000610660.1
|
IFNA13
|
interferon alpha 13
|
|
chr2_-_88947820
Show fit
|
0.04 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5
|
|
chr6_+_154039102
Show fit
|
0.04 |
ENST00000360422.8
ENST00000330432.12
|
OPRM1
|
opioid receptor mu 1
|
|
chr10_+_52128343
Show fit
|
0.04 |
ENST00000672084.1
|
PRKG1
|
protein kinase cGMP-dependent 1
|
|
chr4_+_95091462
Show fit
|
0.04 |
ENST00000264568.8
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr9_-_21166660
Show fit
|
0.04 |
ENST00000380225.1
|
IFNA21
|
interferon alpha 21
|
|
chr16_+_3443645
Show fit
|
0.04 |
ENST00000572757.5
ENST00000573593.5
ENST00000570372.5
ENST00000407558.9
ENST00000424546.6
ENST00000575733.5
ENST00000649205.1
ENST00000574950.5
ENST00000573580.5
ENST00000572169.6
ENST00000649360.1
|
NAA60
|
N-alpha-acetyltransferase 60, NatF catalytic subunit
|
|
chr1_-_149812359
Show fit
|
0.04 |
ENST00000369167.2
ENST00000545683.1
|
H2BC18
|
H2B clustered histone 18
|
|
chr13_+_108629605
Show fit
|
0.04 |
ENST00000457511.7
|
MYO16
|
myosin XVI
|
|
chr3_-_112975018
Show fit
|
0.04 |
ENST00000471858.5
ENST00000308611.8
ENST00000295863.4
|
CD200R1
|
CD200 receptor 1
|
|
chr18_-_3219961
Show fit
|
0.04 |
ENST00000356443.9
|
MYOM1
|
myomesin 1
|
|
chr8_-_7430348
Show fit
|
0.04 |
ENST00000318124.3
|
DEFB103B
|
defensin beta 103B
|
|
chr2_+_233195433
Show fit
|
0.04 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D
|
|
chr19_+_4153616
Show fit
|
0.04 |
ENST00000078445.7
ENST00000595923.5
ENST00000602257.5
ENST00000602147.1
|
CREB3L3
|
cAMP responsive element binding protein 3 like 3
|
|
chr1_-_157820113
Show fit
|
0.04 |
ENST00000368176.8
|
FCRL1
|
Fc receptor like 1
|
|
chr12_+_101594849
Show fit
|
0.04 |
ENST00000547405.5
ENST00000452455.6
ENST00000392934.7
ENST00000547509.5
ENST00000361685.6
ENST00000549145.5
ENST00000361466.7
ENST00000553190.5
ENST00000545503.6
ENST00000536007.5
ENST00000541119.5
ENST00000551300.5
ENST00000550270.1
|
MYBPC1
|
myosin binding protein C1
|
|
chr10_-_28282086
Show fit
|
0.04 |
ENST00000375719.7
ENST00000375732.5
|
MPP7
|
membrane palmitoylated protein 7
|