Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
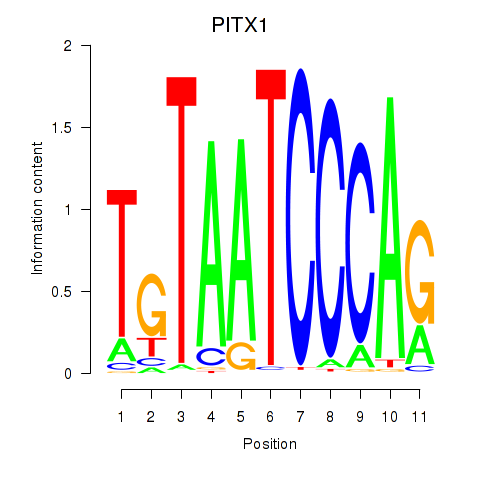
Results for PITX1
Z-value: 1.23
Transcription factors associated with PITX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX1
|
ENSG00000069011.16 | paired like homeodomain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX1 | hg38_v1_chr5_-_135034212_135034294 | 0.70 | 5.2e-02 | Click! |
Activity profile of PITX1 motif
Sorted Z-values of PITX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_6604083 | 0.53 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr3_+_184340337 | 0.47 |
ENST00000310585.4
|
FAM131A
|
family with sequence similarity 131 member A |
| chr19_+_8418386 | 0.45 |
ENST00000602117.1
|
MARCHF2
|
membrane associated ring-CH-type finger 2 |
| chr1_-_161238163 | 0.44 |
ENST00000367982.8
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr2_+_233195433 | 0.40 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr8_+_38404363 | 0.40 |
ENST00000527175.1
|
LETM2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chrX_+_154257538 | 0.38 |
ENST00000599405.3
|
OPN1MW3
|
opsin 1, medium wave sensitive 3 |
| chr1_-_153375591 | 0.37 |
ENST00000368737.5
|
S100A12
|
S100 calcium binding protein A12 |
| chr12_+_104588845 | 0.36 |
ENST00000549016.1
|
CHST11
|
carbohydrate sulfotransferase 11 |
| chr17_+_82559340 | 0.34 |
ENST00000531030.5
ENST00000526383.2 |
FOXK2
|
forkhead box K2 |
| chrX_-_16712865 | 0.33 |
ENST00000380241.7
|
CTPS2
|
CTP synthase 2 |
| chr19_+_38289138 | 0.33 |
ENST00000590738.1
ENST00000587519.4 ENST00000591889.2 |
SPINT2
ENSG00000267748.4
|
serine peptidase inhibitor, Kunitz type 2 novel protein |
| chrX_+_9463272 | 0.33 |
ENST00000407597.7
ENST00000380961.5 ENST00000424279.6 |
TBL1X
|
transducin beta like 1 X-linked |
| chrX_+_3041471 | 0.32 |
ENST00000381127.6
|
ARSF
|
arylsulfatase F |
| chr20_-_32657362 | 0.32 |
ENST00000360785.6
|
C20orf203
|
chromosome 20 open reading frame 203 |
| chr1_+_40247926 | 0.31 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr9_-_127724873 | 0.30 |
ENST00000419060.5
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog |
| chrX_-_2964252 | 0.30 |
ENST00000681963.1
ENST00000684738.1 ENST00000438544.5 ENST00000682184.1 ENST00000545496.6 ENST00000684364.1 ENST00000683677.1 ENST00000683290.1 ENST00000682364.1 |
ARSL
|
arylsulfatase L |
| chr16_+_72008588 | 0.29 |
ENST00000572887.5
ENST00000219240.9 ENST00000574309.5 ENST00000576145.1 |
DHODH
|
dihydroorotate dehydrogenase (quinone) |
| chr5_-_178187364 | 0.29 |
ENST00000463439.3
|
GMCL2
|
germ cell-less 2, spermatogenesis associated |
| chr2_-_171066936 | 0.28 |
ENST00000453628.1
ENST00000434911.6 |
TLK1
|
tousled like kinase 1 |
| chr1_+_28505943 | 0.28 |
ENST00000398958.6
ENST00000649185.1 ENST00000427469.5 ENST00000683442.1 ENST00000434290.6 ENST00000373833.10 ENST00000413987.1 |
RCC1
SNHG3
|
regulator of chromosome condensation 1 small nucleolar RNA host gene 3 |
| chr6_-_73452124 | 0.28 |
ENST00000680833.1
|
CGAS
|
cyclic GMP-AMP synthase |
| chr9_-_111328520 | 0.28 |
ENST00000374428.1
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr19_+_15793951 | 0.28 |
ENST00000308940.8
|
OR10H5
|
olfactory receptor family 10 subfamily H member 5 |
| chr14_+_22495890 | 0.28 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr12_-_21604840 | 0.27 |
ENST00000261195.3
|
GYS2
|
glycogen synthase 2 |
| chr14_+_73591873 | 0.27 |
ENST00000326303.5
|
ACOT4
|
acyl-CoA thioesterase 4 |
| chr19_+_7049321 | 0.27 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3 like 2 |
| chr19_-_2456924 | 0.26 |
ENST00000325327.4
|
LMNB2
|
lamin B2 |
| chr19_-_49061997 | 0.26 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr6_+_42050513 | 0.26 |
ENST00000372978.7
ENST00000494547.5 ENST00000456846.6 ENST00000372982.8 ENST00000472818.5 ENST00000372977.8 |
TAF8
|
TATA-box binding protein associated factor 8 |
| chrX_+_154182596 | 0.26 |
ENST00000595290.6
|
OPN1MW
|
opsin 1, medium wave sensitive |
| chr12_+_53442555 | 0.26 |
ENST00000549135.1
|
PRR13
|
proline rich 13 |
| chr19_-_10577231 | 0.26 |
ENST00000589348.1
ENST00000592285.1 ENST00000587069.5 |
AP1M2
|
adaptor related protein complex 1 subunit mu 2 |
| chr19_-_55141889 | 0.25 |
ENST00000593194.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr19_-_7021431 | 0.25 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr1_+_11664191 | 0.25 |
ENST00000376753.9
|
FBXO6
|
F-box protein 6 |
| chr5_-_157059109 | 0.25 |
ENST00000523175.6
ENST00000522693.5 |
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chrX_+_154219756 | 0.24 |
ENST00000369929.8
|
OPN1MW2
|
opsin 1, medium wave sensitive 2 |
| chr8_-_28083920 | 0.24 |
ENST00000413272.7
|
NUGGC
|
nuclear GTPase, germinal center associated |
| chr19_+_15944299 | 0.24 |
ENST00000641275.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr19_-_12957198 | 0.24 |
ENST00000316939.3
|
GADD45GIP1
|
GADD45G interacting protein 1 |
| chr7_-_76618300 | 0.24 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr10_-_68471911 | 0.23 |
ENST00000358410.8
ENST00000399180.3 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr9_-_113376973 | 0.23 |
ENST00000374180.4
|
HDHD3
|
haloacid dehalogenase like hydrolase domain containing 3 |
| chr9_-_72060605 | 0.23 |
ENST00000377024.8
ENST00000651200.2 ENST00000652752.1 |
C9orf57
|
chromosome 9 open reading frame 57 |
| chr1_-_161238196 | 0.23 |
ENST00000367983.9
ENST00000506209.5 ENST00000367980.6 ENST00000628566.2 |
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr5_-_157058396 | 0.23 |
ENST00000518745.1
ENST00000339252.7 ENST00000625904.2 |
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr11_-_68751441 | 0.23 |
ENST00000544963.1
ENST00000443940.6 ENST00000255087.10 |
TESMIN
|
testis expressed metallothionein like protein |
| chr9_-_72060590 | 0.23 |
ENST00000652156.1
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr21_-_30487436 | 0.23 |
ENST00000334055.5
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr12_+_51239278 | 0.22 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr7_-_97990196 | 0.22 |
ENST00000257627.4
|
OCM2
|
oncomodulin 2 |
| chrX_-_71106728 | 0.22 |
ENST00000374251.6
|
CXorf65
|
chromosome X open reading frame 65 |
| chr4_+_1974633 | 0.22 |
ENST00000677895.1
|
NSD2
|
nuclear receptor binding SET domain protein 2 |
| chr19_+_49676140 | 0.21 |
ENST00000527382.5
ENST00000528623.5 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr19_-_15232399 | 0.21 |
ENST00000221730.8
|
EPHX3
|
epoxide hydrolase 3 |
| chr6_-_34671918 | 0.21 |
ENST00000374021.1
|
ILRUN
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr19_-_15232943 | 0.21 |
ENST00000435261.5
ENST00000594042.1 |
EPHX3
|
epoxide hydrolase 3 |
| chrX_-_2964328 | 0.21 |
ENST00000381134.9
|
ARSL
|
arylsulfatase L |
| chr17_+_18183052 | 0.20 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr12_-_101830671 | 0.20 |
ENST00000549165.1
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta |
| chr12_-_101830799 | 0.20 |
ENST00000549940.5
ENST00000392919.4 |
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta |
| chr19_+_41114430 | 0.20 |
ENST00000331105.7
|
CYP2F1
|
cytochrome P450 family 2 subfamily F member 1 |
| chr19_+_9178979 | 0.19 |
ENST00000642043.1
ENST00000641288.2 |
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr13_-_19505940 | 0.19 |
ENST00000400103.6
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr16_+_25216943 | 0.19 |
ENST00000219660.6
|
AQP8
|
aquaporin 8 |
| chr19_+_40586774 | 0.19 |
ENST00000594298.5
ENST00000597396.5 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr19_-_45639104 | 0.19 |
ENST00000586770.5
ENST00000591721.5 ENST00000245925.8 ENST00000590043.5 ENST00000589876.5 |
EML2
|
EMAP like 2 |
| chr19_+_22832284 | 0.19 |
ENST00000600766.3
|
ZNF723
|
zinc finger protein 723 |
| chr17_+_68249200 | 0.19 |
ENST00000577985.5
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr22_+_24607602 | 0.19 |
ENST00000447416.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr17_-_39778213 | 0.19 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 |
| chr22_+_35383106 | 0.19 |
ENST00000678411.1
|
HMOX1
|
heme oxygenase 1 |
| chr17_+_43006740 | 0.18 |
ENST00000438323.2
ENST00000415816.7 |
IFI35
|
interferon induced protein 35 |
| chr22_-_41940208 | 0.18 |
ENST00000472374.6
|
CENPM
|
centromere protein M |
| chr19_-_45370384 | 0.18 |
ENST00000485403.6
ENST00000586856.1 ENST00000586131.6 ENST00000391945.10 ENST00000684407.1 ENST00000391944.8 |
ERCC2
|
ERCC excision repair 2, TFIIH core complex helicase subunit |
| chr18_+_63702958 | 0.18 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr17_+_8339837 | 0.18 |
ENST00000328248.7
ENST00000584943.1 |
ODF4
|
outer dense fiber of sperm tails 4 |
| chr10_+_123135938 | 0.18 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr16_-_2264221 | 0.18 |
ENST00000566397.5
|
RNPS1
|
RNA binding protein with serine rich domain 1 |
| chr11_+_57805541 | 0.18 |
ENST00000683201.1
ENST00000683769.1 |
CTNND1
|
catenin delta 1 |
| chr1_-_52921681 | 0.18 |
ENST00000467988.2
ENST00000358358.9 ENST00000371522.9 ENST00000536120.5 |
ECHDC2
|
enoyl-CoA hydratase domain containing 2 |
| chr15_-_89221558 | 0.18 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr19_+_10701431 | 0.18 |
ENST00000250237.10
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase catalytic subunit 1 |
| chr19_+_9185594 | 0.18 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chrX_+_35798791 | 0.18 |
ENST00000399985.1
|
MAGEB16
|
MAGE family member B16 |
| chr19_-_41436439 | 0.18 |
ENST00000594660.5
|
DMAC2
|
distal membrane arm assembly complex 2 |
| chrX_+_48761743 | 0.18 |
ENST00000303227.11
|
GLOD5
|
glyoxalase domain containing 5 |
| chr6_-_26189101 | 0.18 |
ENST00000614247.2
|
H4C4
|
H4 clustered histone 4 |
| chr2_-_197198034 | 0.18 |
ENST00000328737.6
|
ANKRD44
|
ankyrin repeat domain 44 |
| chr19_-_15808126 | 0.17 |
ENST00000334920.3
|
OR10H1
|
olfactory receptor family 10 subfamily H member 1 |
| chr17_+_78214286 | 0.17 |
ENST00000592734.5
ENST00000587746.5 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr3_+_42809439 | 0.17 |
ENST00000422265.6
ENST00000487368.4 |
ACKR2
ENSG00000273328.5
|
atypical chemokine receptor 2 novel transcript |
| chr4_+_105710809 | 0.17 |
ENST00000360505.9
ENST00000510865.5 ENST00000509336.5 |
GSTCD
|
glutathione S-transferase C-terminal domain containing |
| chr17_-_81936775 | 0.17 |
ENST00000584848.5
ENST00000577756.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr1_-_108661055 | 0.17 |
ENST00000370031.5
ENST00000651461.1 ENST00000402983.5 ENST00000420055.1 |
HENMT1
|
HEN methyltransferase 1 |
| chr1_-_150720842 | 0.17 |
ENST00000442853.5
ENST00000368995.8 ENST00000322343.11 ENST00000361824.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr2_+_131527833 | 0.16 |
ENST00000295171.10
ENST00000467992.6 ENST00000409856.8 |
CCDC74A
|
coiled-coil domain containing 74A |
| chr7_+_23246697 | 0.16 |
ENST00000381990.6
ENST00000409458.3 ENST00000647578.1 ENST00000258733.9 |
GPNMB
|
glycoprotein nmb |
| chr12_+_101280093 | 0.16 |
ENST00000261637.5
|
UTP20
|
UTP20 small subunit processome component |
| chr1_-_45542726 | 0.16 |
ENST00000676549.1
|
PRDX1
|
peroxiredoxin 1 |
| chr19_+_6364543 | 0.16 |
ENST00000646643.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr8_+_11769639 | 0.16 |
ENST00000436750.7
|
NEIL2
|
nei like DNA glycosylase 2 |
| chr1_-_150876697 | 0.16 |
ENST00000515192.5
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr3_-_196515315 | 0.16 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr19_-_5567984 | 0.16 |
ENST00000448587.5
|
TINCR
|
TINCR ubiquitin domain containing |
| chrX_-_154014246 | 0.16 |
ENST00000444254.1
|
IRAK1
|
interleukin 1 receptor associated kinase 1 |
| chr9_-_37785039 | 0.16 |
ENST00000327304.10
ENST00000396521.3 |
EXOSC3
|
exosome component 3 |
| chr11_-_67443785 | 0.16 |
ENST00000393893.5
ENST00000545016.2 |
CORO1B
|
coronin 1B |
| chr19_+_48954850 | 0.16 |
ENST00000345358.12
ENST00000539787.2 ENST00000415969.6 ENST00000354470.7 ENST00000506183.5 ENST00000391871.4 ENST00000293288.12 |
BAX
|
BCL2 associated X, apoptosis regulator |
| chr19_-_42427379 | 0.16 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr19_-_19203353 | 0.16 |
ENST00000420605.7
ENST00000544883.5 ENST00000538165.2 ENST00000331552.12 |
NR2C2AP
|
nuclear receptor 2C2 associated protein |
| chr13_+_32315071 | 0.16 |
ENST00000544455.6
|
BRCA2
|
BRCA2 DNA repair associated |
| chr2_-_130144994 | 0.16 |
ENST00000457413.1
ENST00000409128.5 ENST00000441670.1 ENST00000409234.3 ENST00000409943.8 ENST00000392984.7 ENST00000310463.10 |
CCDC74B
|
coiled-coil domain containing 74B |
| chr19_-_38949855 | 0.15 |
ENST00000599996.1
|
ENSG00000269547.1
|
novel protein |
| chr7_+_1688119 | 0.15 |
ENST00000424383.4
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr19_+_8052315 | 0.15 |
ENST00000680646.1
|
CCL25
|
C-C motif chemokine ligand 25 |
| chr11_+_35176575 | 0.15 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr17_-_75131729 | 0.15 |
ENST00000245552.7
ENST00000582170.1 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr19_+_9087061 | 0.15 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chrX_+_41339931 | 0.15 |
ENST00000642424.1
|
DDX3X
|
DEAD-box helicase 3 X-linked |
| chr17_+_78214186 | 0.15 |
ENST00000301633.8
ENST00000350051.8 ENST00000374948.6 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr22_-_30546682 | 0.15 |
ENST00000402034.6
|
SEC14L6
|
SEC14 like lipid binding 6 |
| chr17_+_44846318 | 0.15 |
ENST00000591513.5
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr17_-_58280928 | 0.15 |
ENST00000225275.4
|
MPO
|
myeloperoxidase |
| chr1_+_149899618 | 0.15 |
ENST00000369150.1
|
BOLA1
|
bolA family member 1 |
| chr1_+_13060769 | 0.15 |
ENST00000617807.3
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3 |
| chr19_+_8052335 | 0.15 |
ENST00000680507.1
ENST00000680450.1 ENST00000681526.1 ENST00000680506.1 |
CCL25
|
C-C motif chemokine ligand 25 |
| chr19_-_17821505 | 0.15 |
ENST00000598577.1
ENST00000317306.8 ENST00000379695.5 |
INSL3
|
insulin like 3 |
| chr19_-_38256513 | 0.14 |
ENST00000347262.8
ENST00000591585.1 |
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr2_-_223838022 | 0.14 |
ENST00000444408.1
|
AP1S3
|
adaptor related protein complex 1 subunit sigma 3 |
| chr1_+_3752441 | 0.14 |
ENST00000294600.7
|
CCDC27
|
coiled-coil domain containing 27 |
| chr19_+_17751467 | 0.14 |
ENST00000596536.5
ENST00000593870.5 ENST00000598086.5 ENST00000598932.5 ENST00000595023.5 ENST00000594068.5 ENST00000596507.5 ENST00000595033.5 ENST00000597718.5 |
FCHO1
|
FCH and mu domain containing endocytic adaptor 1 |
| chr19_-_15233432 | 0.14 |
ENST00000602233.5
|
EPHX3
|
epoxide hydrolase 3 |
| chr9_-_111330224 | 0.14 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr19_-_12681840 | 0.14 |
ENST00000210060.12
|
DHPS
|
deoxyhypusine synthase |
| chr1_+_156061142 | 0.14 |
ENST00000361084.10
|
RAB25
|
RAB25, member RAS oncogene family |
| chr19_+_1077394 | 0.14 |
ENST00000590577.2
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr1_+_234373439 | 0.14 |
ENST00000366615.10
ENST00000619305.1 |
COA6
|
cytochrome c oxidase assembly factor 6 |
| chr20_-_41300066 | 0.14 |
ENST00000436099.6
ENST00000309060.7 ENST00000373261.5 ENST00000436440.6 ENST00000560364.5 ENST00000560361.5 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr5_+_90899183 | 0.14 |
ENST00000640815.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr5_+_151212117 | 0.14 |
ENST00000523466.5
|
GM2A
|
GM2 ganglioside activator |
| chr19_+_14583076 | 0.14 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr16_-_25015311 | 0.14 |
ENST00000303665.9
ENST00000455311.6 ENST00000289968.11 |
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr17_-_31321743 | 0.14 |
ENST00000247270.3
|
EVI2A
|
ecotropic viral integration site 2A |
| chr19_-_12681771 | 0.14 |
ENST00000351660.9
ENST00000614126.4 |
DHPS
|
deoxyhypusine synthase |
| chr1_-_113887375 | 0.14 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2 like 15 |
| chr10_-_70382589 | 0.14 |
ENST00000373224.5
ENST00000446961.4 ENST00000357631.6 ENST00000358141.6 ENST00000395010.5 |
LRRC20
|
leucine rich repeat containing 20 |
| chr22_+_24607658 | 0.13 |
ENST00000451366.5
ENST00000428855.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr11_+_108116688 | 0.13 |
ENST00000672284.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr8_+_48008409 | 0.13 |
ENST00000523432.5
ENST00000521346.5 ENST00000523111.7 ENST00000517630.5 |
UBE2V2
|
ubiquitin conjugating enzyme E2 V2 |
| chr19_+_48465837 | 0.13 |
ENST00000595676.1
|
ENSG00000268465.1
|
novel protein |
| chrX_+_96883908 | 0.13 |
ENST00000373040.4
|
RPA4
|
replication protein A4 |
| chr1_+_151036578 | 0.13 |
ENST00000368931.8
ENST00000295294.11 |
BNIPL
|
BCL2 interacting protein like |
| chr2_+_130343137 | 0.13 |
ENST00000452955.1
|
IMP4
|
IMP U3 small nucleolar ribonucleoprotein 4 |
| chr12_-_7873027 | 0.13 |
ENST00000542782.5
ENST00000396589.6 ENST00000535266.5 ENST00000542505.5 |
SLC2A14
|
solute carrier family 2 member 14 |
| chr22_-_41947087 | 0.13 |
ENST00000407253.7
ENST00000215980.10 |
CENPM
|
centromere protein M |
| chr12_-_31792290 | 0.13 |
ENST00000340398.5
|
H3-5
|
H3.5 histone |
| chr15_+_45023137 | 0.13 |
ENST00000674211.1
ENST00000267814.14 |
SORD
|
sorbitol dehydrogenase |
| chr16_-_14694308 | 0.13 |
ENST00000438167.8
|
PLA2G10
|
phospholipase A2 group X |
| chr11_+_18266254 | 0.13 |
ENST00000532858.5
ENST00000649195.1 ENST00000356524.9 ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr7_+_134527560 | 0.13 |
ENST00000359579.5
|
AKR1B10
|
aldo-keto reductase family 1 member B10 |
| chr6_-_32371912 | 0.13 |
ENST00000612031.4
|
TSBP1
|
testis expressed basic protein 1 |
| chr16_+_27066919 | 0.13 |
ENST00000505035.3
|
C16orf82
|
chromosome 16 open reading frame 82 |
| chr9_-_122913299 | 0.13 |
ENST00000373659.4
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr6_-_73395133 | 0.13 |
ENST00000441145.1
|
OOEP
|
oocyte expressed protein |
| chr20_+_1113257 | 0.13 |
ENST00000333082.7
ENST00000381899.8 |
PSMF1
|
proteasome inhibitor subunit 1 |
| chr6_+_31706866 | 0.13 |
ENST00000375832.5
ENST00000503322.1 |
LY6G6F
LY6G6F-LY6G6D
|
lymphocyte antigen 6 family member G6F LY6G6F-LY6G6D readthrough |
| chr21_-_38121331 | 0.13 |
ENST00000482032.1
ENST00000398948.5 ENST00000328264.7 ENST00000645093.1 |
DSCR4
KCNJ6
|
Down syndrome critical region 4 potassium inwardly rectifying channel subfamily J member 6 |
| chr7_+_55951852 | 0.13 |
ENST00000285298.9
|
MRPS17
|
mitochondrial ribosomal protein S17 |
| chr11_+_60429567 | 0.13 |
ENST00000300190.7
|
MS4A5
|
membrane spanning 4-domains A5 |
| chr17_-_8210565 | 0.13 |
ENST00000577833.5
ENST00000585124.6 ENST00000534871.5 ENST00000583915.1 ENST00000316199.10 ENST00000581511.5 |
AURKB
|
aurora kinase B |
| chr6_-_73225465 | 0.13 |
ENST00000370388.4
|
KHDC1L
|
KH domain containing 1 like |
| chr19_+_6135635 | 0.13 |
ENST00000588304.5
ENST00000588722.5 ENST00000588485.6 ENST00000591403.5 ENST00000586696.5 ENST00000681525.1 ENST00000589401.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr11_-_69675367 | 0.13 |
ENST00000542341.1
|
LTO1
|
LTO1 maturation factor of ABCE1 |
| chr1_+_13303539 | 0.12 |
ENST00000437300.2
|
PRAMEF33
|
PRAME family member 33 |
| chr6_-_31806937 | 0.12 |
ENST00000375661.6
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr16_+_19067639 | 0.12 |
ENST00000568985.5
ENST00000566110.5 |
COQ7
|
coenzyme Q7, hydroxylase |
| chr7_+_140404034 | 0.12 |
ENST00000537763.6
|
RAB19
|
RAB19, member RAS oncogene family |
| chr7_+_55887277 | 0.12 |
ENST00000426595.1
|
ENSG00000249773.3
|
novel zinc finger protein 713 (ZNF713) and mitochondrial ribosomal protein S17 (MRPS17) protein |
| chr12_-_610407 | 0.12 |
ENST00000397265.7
|
NINJ2
|
ninjurin 2 |
| chr2_-_171231314 | 0.12 |
ENST00000521943.5
|
TLK1
|
tousled like kinase 1 |
| chr19_-_57888780 | 0.12 |
ENST00000595048.5
ENST00000600634.5 ENST00000595295.1 ENST00000596604.5 ENST00000597342.5 ENST00000597807.1 |
ZNF814
|
zinc finger protein 814 |
| chr22_+_39399715 | 0.12 |
ENST00000216160.11
ENST00000331454.3 |
TAB1
|
TGF-beta activated kinase 1 (MAP3K7) binding protein 1 |
| chr22_+_20774092 | 0.12 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr2_+_203706475 | 0.12 |
ENST00000374481.7
ENST00000458610.6 |
CD28
|
CD28 molecule |
| chr5_+_126631680 | 0.12 |
ENST00000357147.4
|
TEX43
|
testis expressed 43 |
| chr16_+_30075967 | 0.12 |
ENST00000279387.12
ENST00000562664.5 ENST00000627746.2 ENST00000562222.5 |
PPP4C
|
protein phosphatase 4 catalytic subunit |
| chr7_+_142332182 | 0.12 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr16_-_30012294 | 0.12 |
ENST00000564979.5
ENST00000563378.5 |
DOC2A
|
double C2 domain alpha |
| chr7_-_100436425 | 0.12 |
ENST00000292330.3
|
PPP1R35
|
protein phosphatase 1 regulatory subunit 35 |
| chr3_-_149576203 | 0.12 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr16_+_67159932 | 0.12 |
ENST00000518148.1
ENST00000258200.8 ENST00000519917.5 ENST00000517382.5 ENST00000521920.5 |
FBXL8
|
F-box and leucine rich repeat protein 8 |
| chr2_-_88979016 | 0.12 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr6_+_63211446 | 0.12 |
ENST00000370659.1
|
FKBP1C
|
FKBP prolyl isomerase family member 1C |
| chr4_+_15703057 | 0.12 |
ENST00000265016.9
ENST00000382346.7 |
BST1
|
bone marrow stromal cell antigen 1 |
| chr8_-_28386417 | 0.12 |
ENST00000521185.5
ENST00000520290.5 ENST00000344423.10 |
ZNF395
|
zinc finger protein 395 |
| chr19_+_58544045 | 0.12 |
ENST00000253024.10
ENST00000593582.5 |
TRIM28
|
tripartite motif containing 28 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.4 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.3 | GO:1903576 | 'de novo' UMP biosynthetic process(GO:0044205) response to L-arginine(GO:1903576) |
| 0.1 | 0.4 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.1 | 0.3 | GO:0018013 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0002339 | B cell selection(GO:0002339) |
| 0.1 | 0.3 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 0.3 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.4 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.3 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.2 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.1 | 0.2 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:0072229 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.0 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0016487 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.0 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.0 | 0.1 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.3 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.1 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.2 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.0 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.4 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 | 0.1 | GO:0060545 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 1.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:1902661 | positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.0 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.1 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:2000325 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 0.2 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.0 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.3 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.9 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.2 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.0 | 0.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:1904492 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.0 | 0.1 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.0 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.0 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 1.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |