Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
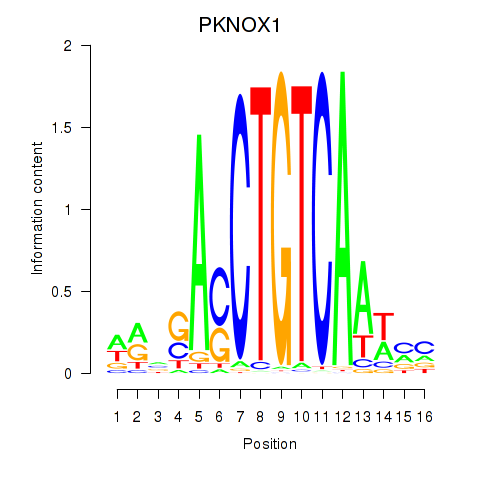
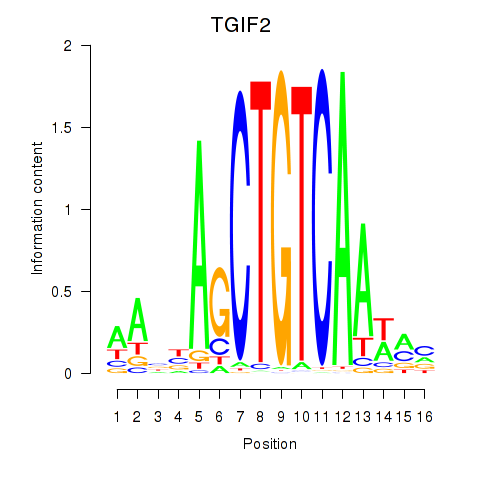
Results for PKNOX1_TGIF2
Z-value: 0.48
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.15 | PBX/knotted 1 homeobox 1 |
|
TGIF2
|
ENSG00000118707.11 | TGFB induced factor homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX1 | hg38_v1_chr21_+_42974510_42974649 | -0.37 | 3.7e-01 | Click! |
| TGIF2 | hg38_v1_chr20_+_36573589_36573723 | 0.30 | 4.7e-01 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_113116676 | 0.35 |
ENST00000671971.1
ENST00000672240.1 ENST00000673240.1 ENST00000673363.1 |
ANK2
|
ankyrin 2 |
| chr12_-_84911178 | 0.23 |
ENST00000681688.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr3_-_190449782 | 0.19 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr8_-_74321532 | 0.17 |
ENST00000342232.5
|
JPH1
|
junctophilin 1 |
| chr1_-_145885826 | 0.17 |
ENST00000544626.2
ENST00000355594.9 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr19_-_38253238 | 0.16 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr16_-_2980406 | 0.16 |
ENST00000431515.6
ENST00000574385.5 ENST00000576268.1 ENST00000574730.5 ENST00000262300.13 ENST00000575632.5 ENST00000573944.5 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr2_-_74553049 | 0.16 |
ENST00000409549.5
|
LOXL3
|
lysyl oxidase like 3 |
| chr21_-_32813679 | 0.15 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr14_-_21526312 | 0.14 |
ENST00000537235.2
|
SALL2
|
spalt like transcription factor 2 |
| chr19_+_9185594 | 0.14 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr14_+_103100328 | 0.14 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3 like 4 |
| chr1_-_54406385 | 0.13 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr8_+_31639222 | 0.13 |
ENST00000519301.6
ENST00000652698.1 |
NRG1
|
neuregulin 1 |
| chr12_-_91058016 | 0.13 |
ENST00000266719.4
|
KERA
|
keratocan |
| chr2_-_74552616 | 0.12 |
ENST00000409249.5
|
LOXL3
|
lysyl oxidase like 3 |
| chr8_+_31639291 | 0.12 |
ENST00000651149.1
ENST00000650866.1 |
NRG1
|
neuregulin 1 |
| chr2_+_105851748 | 0.11 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr21_-_32813695 | 0.11 |
ENST00000479548.2
ENST00000490358.5 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr8_-_123396412 | 0.11 |
ENST00000287394.10
|
ATAD2
|
ATPase family AAA domain containing 2 |
| chr1_-_205422050 | 0.11 |
ENST00000367153.9
|
LEMD1
|
LEM domain containing 1 |
| chr4_+_3074661 | 0.10 |
ENST00000355072.11
|
HTT
|
huntingtin |
| chr18_-_54959391 | 0.09 |
ENST00000591504.6
|
CCDC68
|
coiled-coil domain containing 68 |
| chr3_+_111542178 | 0.09 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr2_+_233060295 | 0.09 |
ENST00000445964.6
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr14_-_100376251 | 0.09 |
ENST00000556645.5
ENST00000556209.5 ENST00000556504.5 ENST00000556435.5 ENST00000554772.5 ENST00000553581.1 ENST00000553769.6 ENST00000554605.5 ENST00000557722.5 ENST00000553413.5 ENST00000553524.5 ENST00000358655.8 |
WARS1
|
tryptophanyl-tRNA synthetase 1 |
| chr3_+_52794768 | 0.09 |
ENST00000621946.4
ENST00000416872.6 ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr1_-_209651291 | 0.09 |
ENST00000391911.5
ENST00000415782.1 |
LAMB3
|
laminin subunit beta 3 |
| chr9_-_33447553 | 0.08 |
ENST00000645858.1
ENST00000297991.6 |
AQP3
|
aquaporin 3 (Gill blood group) |
| chr14_-_105168753 | 0.08 |
ENST00000331782.8
ENST00000347004.2 |
JAG2
|
jagged canonical Notch ligand 2 |
| chr2_+_234050732 | 0.08 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2 |
| chr19_-_48993300 | 0.08 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr21_+_37420299 | 0.08 |
ENST00000455097.6
ENST00000643854.1 ENST00000645424.1 ENST00000642309.1 ENST00000645774.1 ENST00000398956.2 |
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr3_+_57890011 | 0.08 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr12_+_121210160 | 0.08 |
ENST00000542067.5
|
P2RX4
|
purinergic receptor P2X 4 |
| chr20_-_3084716 | 0.08 |
ENST00000380293.3
|
AVP
|
arginine vasopressin |
| chr14_-_94390650 | 0.08 |
ENST00000449399.7
ENST00000404814.8 |
SERPINA1
|
serpin family A member 1 |
| chr12_+_121210065 | 0.08 |
ENST00000359949.11
ENST00000337233.9 ENST00000538701.5 |
P2RX4
|
purinergic receptor P2X 4 |
| chr14_+_61529005 | 0.08 |
ENST00000556347.1
|
ENSG00000258989.1
|
novel protein |
| chr1_+_76867469 | 0.08 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr17_+_80544817 | 0.07 |
ENST00000306801.8
ENST00000570891.5 |
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr16_-_69384747 | 0.07 |
ENST00000566257.5
|
TERF2
|
telomeric repeat binding factor 2 |
| chr1_+_207089283 | 0.07 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr2_+_233760265 | 0.07 |
ENST00000305208.10
ENST00000360418.4 |
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1 |
| chr14_-_21024092 | 0.07 |
ENST00000554398.5
|
NDRG2
|
NDRG family member 2 |
| chr1_+_207089195 | 0.07 |
ENST00000452902.6
|
C4BPB
|
complement component 4 binding protein beta |
| chr6_+_137867414 | 0.07 |
ENST00000237289.8
ENST00000433680.1 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr1_+_205504644 | 0.07 |
ENST00000429964.7
ENST00000443813.6 |
CDK18
|
cyclin dependent kinase 18 |
| chr3_-_183162726 | 0.07 |
ENST00000265598.8
|
LAMP3
|
lysosomal associated membrane protein 3 |
| chr9_-_122227525 | 0.07 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr1_+_207089233 | 0.07 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr1_+_205504592 | 0.07 |
ENST00000506784.5
ENST00000360066.6 |
CDK18
|
cyclin dependent kinase 18 |
| chr14_-_21023954 | 0.07 |
ENST00000554094.5
|
NDRG2
|
NDRG family member 2 |
| chr19_-_12933680 | 0.07 |
ENST00000593021.1
ENST00000314606.9 ENST00000587981.1 ENST00000423140.6 |
FARSA
|
phenylalanyl-tRNA synthetase subunit alpha |
| chr6_+_110874775 | 0.06 |
ENST00000675380.1
ENST00000368882.8 ENST00000368877.9 ENST00000368885.8 ENST00000672937.2 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr1_+_207088825 | 0.06 |
ENST00000367078.8
|
C4BPB
|
complement component 4 binding protein beta |
| chr7_+_141776674 | 0.06 |
ENST00000247881.4
|
TAS2R4
|
taste 2 receptor member 4 |
| chr1_+_156106698 | 0.06 |
ENST00000675667.1
|
LMNA
|
lamin A/C |
| chr20_+_56412249 | 0.06 |
ENST00000679887.1
ENST00000434344.2 |
CASS4
|
Cas scaffold protein family member 4 |
| chr17_+_80545422 | 0.06 |
ENST00000544334.6
|
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr1_-_186680411 | 0.06 |
ENST00000367468.10
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 |
| chr14_-_94390667 | 0.06 |
ENST00000557492.5
ENST00000355814.8 ENST00000437397.5 ENST00000448921.5 ENST00000393088.8 |
SERPINA1
|
serpin family A member 1 |
| chr2_+_62673851 | 0.06 |
ENST00000405015.7
ENST00000413434.5 ENST00000426940.5 ENST00000449820.5 |
EHBP1
|
EH domain binding protein 1 |
| chr5_+_66828762 | 0.06 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_98825628 | 0.06 |
ENST00000617590.1
ENST00000518165.5 ENST00000419617.7 |
STK3
|
serine/threonine kinase 3 |
| chrX_-_15664798 | 0.06 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator |
| chr16_-_57797764 | 0.06 |
ENST00000465878.6
ENST00000561524.5 |
KIFC3
|
kinesin family member C3 |
| chr14_-_94390614 | 0.05 |
ENST00000553327.5
ENST00000556955.5 ENST00000557118.5 ENST00000440909.5 |
SERPINA1
|
serpin family A member 1 |
| chr10_-_11532275 | 0.05 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr20_+_56412393 | 0.05 |
ENST00000679529.1
|
CASS4
|
Cas scaffold protein family member 4 |
| chr20_+_56412112 | 0.05 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4 |
| chr17_-_35795592 | 0.05 |
ENST00000615136.4
ENST00000605424.6 ENST00000612672.1 |
MMP28
|
matrix metallopeptidase 28 |
| chr12_-_31729010 | 0.05 |
ENST00000537562.5
ENST00000537960.5 ENST00000281471.11 ENST00000536761.5 ENST00000542781.5 ENST00000457428.6 |
AMN1
|
antagonist of mitotic exit network 1 homolog |
| chr4_+_2418932 | 0.05 |
ENST00000635017.1
|
CFAP99
|
cilia and flagella associated protein 99 |
| chr1_+_154993581 | 0.05 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr16_+_81645352 | 0.05 |
ENST00000398040.8
|
CMIP
|
c-Maf inducing protein |
| chr16_+_27402167 | 0.05 |
ENST00000564089.5
ENST00000337929.8 |
IL21R
|
interleukin 21 receptor |
| chr4_+_127965429 | 0.05 |
ENST00000513371.1
ENST00000611882.1 |
ABHD18
|
abhydrolase domain containing 18 |
| chr18_+_56651335 | 0.05 |
ENST00000589935.1
ENST00000254442.8 ENST00000357574.7 |
WDR7
|
WD repeat domain 7 |
| chr11_+_129375841 | 0.05 |
ENST00000281437.6
|
BARX2
|
BARX homeobox 2 |
| chr1_+_109213887 | 0.05 |
ENST00000234677.7
ENST00000369923.4 |
SARS1
|
seryl-tRNA synthetase 1 |
| chr22_-_49825900 | 0.05 |
ENST00000404034.5
|
BRD1
|
bromodomain containing 1 |
| chr18_+_56651385 | 0.05 |
ENST00000615645.4
|
WDR7
|
WD repeat domain 7 |
| chr3_-_45796467 | 0.05 |
ENST00000353278.8
ENST00000456124.6 |
SLC6A20
|
solute carrier family 6 member 20 |
| chr12_+_40692413 | 0.05 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr17_+_40140500 | 0.05 |
ENST00000264645.12
|
CASC3
|
CASC3 exon junction complex subunit |
| chr1_-_242449478 | 0.05 |
ENST00000427495.5
|
PLD5
|
phospholipase D family member 5 |
| chr3_-_112133218 | 0.04 |
ENST00000488580.5
ENST00000308910.9 ENST00000460387.6 ENST00000484193.5 ENST00000487901.1 |
GCSAM
|
germinal center associated signaling and motility |
| chr12_-_31326171 | 0.04 |
ENST00000542983.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr16_-_20806431 | 0.04 |
ENST00000357967.9
ENST00000300005.7 ENST00000569729.5 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr17_-_81869934 | 0.04 |
ENST00000580685.5
|
ARHGDIA
|
Rho GDP dissociation inhibitor alpha |
| chr5_-_95284535 | 0.04 |
ENST00000515393.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr7_-_105269007 | 0.04 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2 |
| chr7_+_80135694 | 0.04 |
ENST00000457358.7
|
GNAI1
|
G protein subunit alpha i1 |
| chr19_-_6424802 | 0.04 |
ENST00000600480.2
|
KHSRP
|
KH-type splicing regulatory protein |
| chr19_+_15793951 | 0.04 |
ENST00000308940.8
|
OR10H5
|
olfactory receptor family 10 subfamily H member 5 |
| chr4_+_83536097 | 0.04 |
ENST00000395226.6
ENST00000264409.5 |
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr2_-_71227055 | 0.04 |
ENST00000244221.9
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr2_+_33436304 | 0.04 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr18_-_70205655 | 0.04 |
ENST00000255674.11
ENST00000640769.2 |
RTTN
|
rotatin |
| chr7_+_95485898 | 0.04 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr17_+_70075317 | 0.04 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr3_+_37975773 | 0.04 |
ENST00000436654.1
|
CTDSPL
|
CTD small phosphatase like |
| chrX_+_47078380 | 0.04 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chr6_+_137867241 | 0.04 |
ENST00000612899.5
ENST00000420009.5 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr3_-_27722699 | 0.04 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr3_-_142448060 | 0.04 |
ENST00000264951.8
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr12_-_21604840 | 0.04 |
ENST00000261195.3
|
GYS2
|
glycogen synthase 2 |
| chr2_+_234050679 | 0.04 |
ENST00000373368.5
ENST00000168148.8 |
SPP2
|
secreted phosphoprotein 2 |
| chr11_-_46846233 | 0.04 |
ENST00000529230.6
ENST00000312055.9 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr10_+_92848461 | 0.04 |
ENST00000443748.6
ENST00000371543.5 ENST00000260762.10 |
EXOC6
|
exocyst complex component 6 |
| chr11_+_64291992 | 0.04 |
ENST00000394525.6
|
KCNK4
|
potassium two pore domain channel subfamily K member 4 |
| chr1_-_246566238 | 0.04 |
ENST00000366514.5
|
TFB2M
|
transcription factor B2, mitochondrial |
| chr11_-_59615673 | 0.03 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein |
| chr14_-_21526391 | 0.03 |
ENST00000611430.4
|
SALL2
|
spalt like transcription factor 2 |
| chr11_+_125746272 | 0.03 |
ENST00000305738.10
ENST00000437148.2 |
PATE1
|
prostate and testis expressed 1 |
| chr17_+_70075215 | 0.03 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr12_+_62260374 | 0.03 |
ENST00000312635.10
ENST00000280377.10 ENST00000549237.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr5_+_142771119 | 0.03 |
ENST00000642734.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr8_-_20303955 | 0.03 |
ENST00000381569.5
|
LZTS1
|
leucine zipper tumor suppressor 1 |
| chr22_-_29838227 | 0.03 |
ENST00000307790.8
ENST00000397771.6 ENST00000542393.5 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr19_+_48993525 | 0.03 |
ENST00000601968.5
ENST00000596837.5 |
RUVBL2
|
RuvB like AAA ATPase 2 |
| chr12_-_31326142 | 0.03 |
ENST00000337682.9
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr1_-_63523175 | 0.03 |
ENST00000371092.7
ENST00000271002.15 |
ITGB3BP
|
integrin subunit beta 3 binding protein |
| chr19_-_6424772 | 0.03 |
ENST00000619396.4
ENST00000398148.7 |
KHSRP
|
KH-type splicing regulatory protein |
| chr3_-_33440343 | 0.03 |
ENST00000283629.8
|
UBP1
|
upstream binding protein 1 |
| chr12_-_31326111 | 0.03 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr11_+_827545 | 0.03 |
ENST00000528542.6
|
CRACR2B
|
calcium release activated channel regulator 2B |
| chr2_+_190343561 | 0.03 |
ENST00000322522.8
ENST00000392329.7 ENST00000430311.5 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr18_+_34493386 | 0.03 |
ENST00000679936.1
|
DTNA
|
dystrobrevin alpha |
| chr3_-_27722316 | 0.03 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr11_+_827913 | 0.03 |
ENST00000525077.2
|
CRACR2B
|
calcium release activated channel regulator 2B |
| chr9_-_120842898 | 0.03 |
ENST00000625444.1
ENST00000210313.8 ENST00000373904.5 |
PSMD5
|
proteasome 26S subunit, non-ATPase 5 |
| chr19_+_48872412 | 0.03 |
ENST00000200453.6
|
PPP1R15A
|
protein phosphatase 1 regulatory subunit 15A |
| chr2_+_99337364 | 0.03 |
ENST00000617677.1
ENST00000289371.11 |
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr8_-_17697654 | 0.03 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr1_-_44141631 | 0.03 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chrX_+_16786421 | 0.03 |
ENST00000398155.4
ENST00000380122.10 |
TXLNG
|
taxilin gamma |
| chr4_+_56907876 | 0.03 |
ENST00000640168.2
ENST00000309042.12 |
REST
|
RE1 silencing transcription factor |
| chr6_+_54307856 | 0.03 |
ENST00000370869.7
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr12_+_96912517 | 0.03 |
ENST00000457368.2
|
NEDD1
|
NEDD1 gamma-tubulin ring complex targeting factor |
| chr19_+_7669080 | 0.03 |
ENST00000629642.1
|
RETN
|
resistin |
| chr2_+_112275588 | 0.03 |
ENST00000409871.6
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr7_+_64045420 | 0.03 |
ENST00000647787.1
ENST00000456806.3 |
ENSG00000285544.1
ZNF727
|
novel transcript zinc finger protein 727 |
| chr1_+_110034607 | 0.03 |
ENST00000369795.8
|
STRIP1
|
striatin interacting protein 1 |
| chr18_+_34493428 | 0.03 |
ENST00000682483.1
|
DTNA
|
dystrobrevin alpha |
| chrX_-_135850791 | 0.03 |
ENST00000611438.1
|
CT45A8
|
cancer/testis antigen family 45 member A8 |
| chr1_-_247008042 | 0.03 |
ENST00000339986.8
ENST00000487338.6 |
ZNF695
|
zinc finger protein 695 |
| chr8_-_18684093 | 0.03 |
ENST00000428502.6
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_+_130579556 | 0.03 |
ENST00000319725.10
|
FUBP3
|
far upstream element binding protein 3 |
| chr13_+_24270681 | 0.03 |
ENST00000343003.10
ENST00000399949.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr6_-_162727748 | 0.03 |
ENST00000366892.5
ENST00000366898.6 ENST00000366897.5 ENST00000366896.5 ENST00000674250.1 |
PRKN
|
parkin RBR E3 ubiquitin protein ligase |
| chr12_+_6852115 | 0.03 |
ENST00000389231.9
ENST00000229268.13 ENST00000542087.1 |
USP5
|
ubiquitin specific peptidase 5 |
| chr6_-_32173109 | 0.03 |
ENST00000395496.5
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr13_-_46142834 | 0.03 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr16_+_30201057 | 0.02 |
ENST00000569485.5
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr7_-_42237187 | 0.02 |
ENST00000395925.8
|
GLI3
|
GLI family zinc finger 3 |
| chrX_-_135868069 | 0.02 |
ENST00000604569.1
|
CT45A9
|
cancer/testis antigen family 45 member A9 |
| chr8_-_18684033 | 0.02 |
ENST00000614430.3
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr3_-_45796518 | 0.02 |
ENST00000413781.1
ENST00000358525.9 |
SLC6A20
|
solute carrier family 6 member 20 |
| chr11_-_107858777 | 0.02 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr9_-_128191878 | 0.02 |
ENST00000538431.5
|
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr15_-_30991595 | 0.02 |
ENST00000435680.6
|
MTMR10
|
myotubularin related protein 10 |
| chr3_+_49689531 | 0.02 |
ENST00000432042.5
ENST00000454491.5 ENST00000327697.11 |
RNF123
|
ring finger protein 123 |
| chr7_+_74964692 | 0.02 |
ENST00000616305.2
|
CASTOR2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr22_-_37244417 | 0.02 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chr12_+_121888751 | 0.02 |
ENST00000261817.6
ENST00000538613.5 ENST00000542602.1 ENST00000541212.6 |
PSMD9
|
proteasome 26S subunit, non-ATPase 9 |
| chr11_+_828150 | 0.02 |
ENST00000450448.5
|
CRACR2B
|
calcium release activated channel regulator 2B |
| chrX_-_135816265 | 0.02 |
ENST00000605791.7
|
CT45A2
|
cancer/testis antigen family 45 member A2 |
| chr6_+_30328889 | 0.02 |
ENST00000396547.5
ENST00000623385.3 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr6_+_167122742 | 0.02 |
ENST00000341935.9
ENST00000349984.6 |
CCR6
|
C-C motif chemokine receptor 6 |
| chr16_+_21233672 | 0.02 |
ENST00000311620.7
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr10_-_72088972 | 0.02 |
ENST00000317376.8
ENST00000412663.5 |
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr3_-_142448028 | 0.02 |
ENST00000392981.7
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr17_-_3595831 | 0.02 |
ENST00000399759.7
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chr19_+_48993864 | 0.02 |
ENST00000595090.6
|
RUVBL2
|
RuvB like AAA ATPase 2 |
| chr16_-_58295019 | 0.02 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr19_+_10718114 | 0.02 |
ENST00000408974.8
|
DNM2
|
dynamin 2 |
| chrX_+_102125703 | 0.02 |
ENST00000329035.2
|
TCEAL2
|
transcription elongation factor A like 2 |
| chrX_+_102125668 | 0.02 |
ENST00000372780.6
|
TCEAL2
|
transcription elongation factor A like 2 |
| chr7_+_129375643 | 0.02 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr11_-_101129706 | 0.02 |
ENST00000534013.5
|
PGR
|
progesterone receptor |
| chr8_-_18683932 | 0.02 |
ENST00000615573.4
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr12_-_6851245 | 0.02 |
ENST00000540683.1
ENST00000229265.10 ENST00000535406.5 ENST00000422785.7 ENST00000538862.7 |
CDCA3
|
cell division cycle associated 3 |
| chr7_+_76510528 | 0.02 |
ENST00000334348.8
|
UPK3B
|
uroplakin 3B |
| chr19_+_10718047 | 0.02 |
ENST00000585892.5
ENST00000355667.11 ENST00000389253.9 ENST00000359692.10 |
DNM2
|
dynamin 2 |
| chr10_-_97292625 | 0.02 |
ENST00000466484.1
ENST00000358531.9 ENST00000358308.7 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr16_-_58294976 | 0.02 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chr6_-_100881281 | 0.02 |
ENST00000369143.2
ENST00000369162.7 ENST00000324723.10 ENST00000522650.5 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr7_+_76510516 | 0.02 |
ENST00000257632.9
|
UPK3B
|
uroplakin 3B |
| chr10_-_117005349 | 0.02 |
ENST00000615301.4
|
SHTN1
|
shootin 1 |
| chr10_-_117005570 | 0.02 |
ENST00000260777.14
ENST00000392903.3 |
SHTN1
|
shootin 1 |
| chr17_-_31901658 | 0.02 |
ENST00000261708.9
|
UTP6
|
UTP6 small subunit processome component |
| chr16_-_70251894 | 0.02 |
ENST00000435634.3
|
EXOSC6
|
exosome component 6 |
| chr4_+_71062642 | 0.02 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr2_-_196926717 | 0.02 |
ENST00000409475.5
ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr4_+_158210479 | 0.02 |
ENST00000504569.5
ENST00000509278.5 ENST00000514558.5 ENST00000503200.5 ENST00000296529.11 |
TMEM144
|
transmembrane protein 144 |
| chrX_-_135764444 | 0.02 |
ENST00000597510.6
|
CT45A3
|
cancer/testis antigen family 45 member A3 |
| chr9_-_131740056 | 0.02 |
ENST00000372195.5
ENST00000683357.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor 1 |
| chr11_-_2903490 | 0.02 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:1903824 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.0 | GO:0061573 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.2 | GO:0032308 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.0 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |