Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
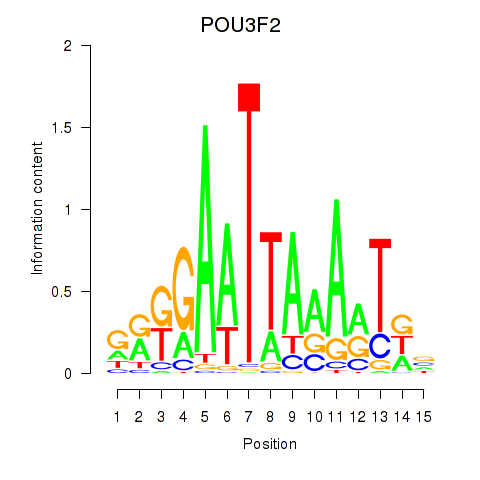
Results for POU3F2
Z-value: 0.79
Transcription factors associated with POU3F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F2
|
ENSG00000184486.10 | POU class 3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU3F2 | hg38_v1_chr6_+_98834560_98834582 | 0.57 | 1.4e-01 | Click! |
Activity profile of POU3F2 motif
Sorted Z-values of POU3F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_63887698 | 1.18 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr5_+_140848360 | 0.88 |
ENST00000532602.2
|
PCDHA9
|
protocadherin alpha 9 |
| chr11_-_118252279 | 0.51 |
ENST00000525386.5
ENST00000527472.1 ENST00000278949.9 |
MPZL3
|
myelin protein zero like 3 |
| chr18_+_31447732 | 0.50 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr8_-_15238423 | 0.49 |
ENST00000382080.6
|
SGCZ
|
sarcoglycan zeta |
| chr1_-_242449478 | 0.49 |
ENST00000427495.5
|
PLD5
|
phospholipase D family member 5 |
| chr2_-_112784486 | 0.47 |
ENST00000263339.4
|
IL1A
|
interleukin 1 alpha |
| chr5_+_140834230 | 0.44 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr1_+_101238090 | 0.42 |
ENST00000475289.2
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr4_-_152679984 | 0.41 |
ENST00000304385.8
ENST00000504064.1 |
TMEM154
|
transmembrane protein 154 |
| chr6_+_54846735 | 0.40 |
ENST00000306858.8
|
FAM83B
|
family with sequence similarity 83 member B |
| chr4_-_15938740 | 0.39 |
ENST00000382333.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr8_-_85341705 | 0.39 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr2_-_112836702 | 0.36 |
ENST00000416750.1
ENST00000263341.7 ENST00000418817.5 |
IL1B
|
interleukin 1 beta |
| chr18_+_63777773 | 0.36 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr18_+_63775395 | 0.36 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr5_+_140786136 | 0.35 |
ENST00000378133.4
ENST00000504120.4 |
PCDHA1
|
protocadherin alpha 1 |
| chr18_+_23949847 | 0.35 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3 |
| chr6_+_106086316 | 0.34 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr19_+_35115912 | 0.34 |
ENST00000603181.5
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr8_-_6978634 | 0.33 |
ENST00000382679.2
|
DEFA1
|
defensin alpha 1 |
| chrX_+_106693838 | 0.33 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr13_-_20230970 | 0.32 |
ENST00000644667.1
ENST00000646108.1 |
GJB6
|
gap junction protein beta 6 |
| chr15_+_43593054 | 0.32 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr2_-_216013582 | 0.32 |
ENST00000620139.4
|
MREG
|
melanoregulin |
| chr11_-_5301946 | 0.32 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr19_-_6720641 | 0.31 |
ENST00000245907.11
|
C3
|
complement C3 |
| chr5_+_140841183 | 0.30 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr11_-_102798148 | 0.30 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1 |
| chr20_+_59721210 | 0.30 |
ENST00000395636.6
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_-_71784046 | 0.30 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr12_-_11134644 | 0.30 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr1_+_31576485 | 0.30 |
ENST00000457433.6
ENST00000271064.12 |
TINAGL1
|
tubulointerstitial nephritis antigen like 1 |
| chr18_+_63775369 | 0.29 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr6_+_47656436 | 0.29 |
ENST00000507065.5
ENST00000296862.5 |
ADGRF2
|
adhesion G protein-coupled receptor F2 |
| chr6_+_30888672 | 0.29 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_+_116427839 | 0.29 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr18_+_63752935 | 0.29 |
ENST00000425392.5
ENST00000336429.6 |
SERPINB7
|
serpin family B member 7 |
| chr12_-_11022620 | 0.28 |
ENST00000390673.2
|
TAS2R19
|
taste 2 receptor member 19 |
| chr15_-_78944985 | 0.28 |
ENST00000615999.5
ENST00000677789.1 ENST00000676880.1 ENST00000677936.1 ENST00000220166.10 ENST00000677810.1 ENST00000678644.1 ENST00000677534.1 ENST00000677316.1 |
CTSH
|
cathepsin H |
| chr18_+_49562049 | 0.28 |
ENST00000261292.9
ENST00000427224.6 ENST00000580036.5 |
LIPG
|
lipase G, endothelial type |
| chr9_+_21409116 | 0.28 |
ENST00000380205.2
|
IFNA8
|
interferon alpha 8 |
| chr21_+_38272410 | 0.28 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr1_+_86547070 | 0.28 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr12_-_11062294 | 0.28 |
ENST00000533467.1
|
TAS2R46
|
taste 2 receptor member 46 |
| chr12_-_84892120 | 0.27 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr6_+_29306626 | 0.26 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr8_+_18391276 | 0.26 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr19_+_35115872 | 0.26 |
ENST00000435734.6
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr18_+_23873000 | 0.26 |
ENST00000269217.11
ENST00000587184.5 |
LAMA3
|
laminin subunit alpha 3 |
| chr7_-_20217342 | 0.25 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr12_+_8822610 | 0.25 |
ENST00000299698.12
|
A2ML1
|
alpha-2-macroglobulin like 1 |
| chr17_-_41521719 | 0.25 |
ENST00000393976.6
|
KRT15
|
keratin 15 |
| chrX_+_66164210 | 0.25 |
ENST00000343002.7
ENST00000336279.9 |
HEPH
|
hephaestin |
| chr19_-_43204223 | 0.24 |
ENST00000599746.5
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr4_+_40197023 | 0.24 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr13_+_73054969 | 0.23 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5 |
| chr2_-_216013517 | 0.23 |
ENST00000263268.11
|
MREG
|
melanoregulin |
| chr3_+_187024614 | 0.23 |
ENST00000416235.6
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr5_-_160312756 | 0.23 |
ENST00000644313.1
|
CCNJL
|
cyclin J like |
| chr5_+_140827950 | 0.23 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr9_-_39239174 | 0.23 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr19_-_50953063 | 0.23 |
ENST00000391809.6
|
KLK5
|
kallikrein related peptidase 5 |
| chr8_+_49911801 | 0.23 |
ENST00000643809.1
|
SNTG1
|
syntrophin gamma 1 |
| chr4_-_56821679 | 0.23 |
ENST00000504762.1
ENST00000506738.6 ENST00000248701.8 ENST00000616980.1 ENST00000618802.3 ENST00000631082.1 |
SPINK2
|
serine peptidase inhibitor Kazal type 2 |
| chr6_+_29301701 | 0.23 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr19_-_50953093 | 0.22 |
ENST00000593428.5
|
KLK5
|
kallikrein related peptidase 5 |
| chr20_+_59835853 | 0.22 |
ENST00000492611.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr19_-_10587219 | 0.22 |
ENST00000591240.5
ENST00000589684.5 ENST00000591676.1 ENST00000250244.11 ENST00000590923.5 |
AP1M2
|
adaptor related protein complex 1 subunit mu 2 |
| chr4_+_87975667 | 0.22 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr13_+_31739542 | 0.22 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr17_-_4739866 | 0.22 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chr18_+_63587297 | 0.22 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr18_+_63587336 | 0.22 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13 |
| chr3_-_151329539 | 0.22 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr20_-_57711536 | 0.22 |
ENST00000265626.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_+_68734773 | 0.22 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr19_-_50952942 | 0.22 |
ENST00000594846.1
ENST00000336334.8 |
KLK5
|
kallikrein related peptidase 5 |
| chr1_-_207032749 | 0.21 |
ENST00000359470.6
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr6_-_136466858 | 0.21 |
ENST00000544465.5
|
MAP7
|
microtubule associated protein 7 |
| chr12_-_70637405 | 0.21 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr7_-_80922354 | 0.21 |
ENST00000419255.6
|
SEMA3C
|
semaphorin 3C |
| chr13_+_31739520 | 0.21 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr12_+_50924005 | 0.21 |
ENST00000550502.1
|
METTL7A
|
methyltransferase like 7A |
| chr6_-_47042306 | 0.20 |
ENST00000371253.7
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr11_-_55936400 | 0.20 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr5_+_140875299 | 0.20 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chr7_+_148133684 | 0.20 |
ENST00000628930.2
|
CNTNAP2
|
contactin associated protein 2 |
| chr5_+_148202771 | 0.20 |
ENST00000514389.5
ENST00000621437.4 |
SPINK6
|
serine peptidase inhibitor Kazal type 6 |
| chr4_+_40196907 | 0.20 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr18_-_31102411 | 0.20 |
ENST00000251081.8
ENST00000280904.11 ENST00000682357.1 ENST00000648081.1 |
DSC2
|
desmocollin 2 |
| chr19_-_51028015 | 0.20 |
ENST00000319720.11
|
KLK11
|
kallikrein related peptidase 11 |
| chr11_-_102780620 | 0.20 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr3_-_172523460 | 0.20 |
ENST00000420541.6
|
TNFSF10
|
TNF superfamily member 10 |
| chr9_-_5833014 | 0.19 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr15_+_94297939 | 0.19 |
ENST00000357742.9
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chrX_-_31178149 | 0.19 |
ENST00000679437.1
|
DMD
|
dystrophin |
| chr2_+_203936755 | 0.19 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr5_+_35856883 | 0.19 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr14_+_21280077 | 0.19 |
ENST00000400017.7
|
RPGRIP1
|
RPGR interacting protein 1 |
| chr11_-_112164056 | 0.19 |
ENST00000524595.5
|
IL18
|
interleukin 18 |
| chr11_+_18132565 | 0.19 |
ENST00000621697.2
|
MRGPRX3
|
MAS related GPR family member X3 |
| chr11_-_112164080 | 0.19 |
ENST00000528832.1
ENST00000280357.12 |
IL18
|
interleukin 18 |
| chr5_-_16508812 | 0.19 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr6_-_32816910 | 0.19 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chrX_+_106168297 | 0.19 |
ENST00000337685.6
ENST00000357175.6 |
PWWP3B
|
PWWP domain containing 3B |
| chrX_-_54798253 | 0.19 |
ENST00000218436.7
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family member 6 |
| chr1_+_61952283 | 0.19 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr4_+_87975829 | 0.19 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_61952036 | 0.19 |
ENST00000646453.1
ENST00000635137.1 |
PATJ
|
PATJ crumbs cell polarity complex component |
| chr6_+_36130484 | 0.19 |
ENST00000373766.9
ENST00000211287.9 |
MAPK13
|
mitogen-activated protein kinase 13 |
| chr16_+_4795357 | 0.18 |
ENST00000586005.6
|
SMIM22
|
small integral membrane protein 22 |
| chr8_+_31639291 | 0.18 |
ENST00000651149.1
ENST00000650866.1 |
NRG1
|
neuregulin 1 |
| chr19_-_42594918 | 0.18 |
ENST00000244336.10
|
CEACAM8
|
CEA cell adhesion molecule 8 |
| chr4_-_88158605 | 0.18 |
ENST00000237612.8
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr6_-_26216673 | 0.18 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chrX_+_3066833 | 0.18 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr5_-_177780845 | 0.18 |
ENST00000393518.7
|
FAM153A
|
family with sequence similarity 153 member A |
| chr1_+_61742418 | 0.18 |
ENST00000316485.11
ENST00000371158.6 ENST00000642238.2 ENST00000613764.4 |
PATJ
|
PATJ crumbs cell polarity complex component |
| chr7_-_22194709 | 0.18 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr8_+_31639222 | 0.18 |
ENST00000519301.6
ENST00000652698.1 |
NRG1
|
neuregulin 1 |
| chr2_+_218710931 | 0.18 |
ENST00000442769.5
ENST00000424644.1 |
TTLL4
|
tubulin tyrosine ligase like 4 |
| chr17_-_41118369 | 0.18 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr1_+_34782259 | 0.18 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chr19_-_6767420 | 0.18 |
ENST00000245908.11
ENST00000437152.7 ENST00000597687.1 |
SH2D3A
|
SH2 domain containing 3A |
| chr18_+_63476927 | 0.18 |
ENST00000489441.5
ENST00000382771.9 ENST00000424602.1 |
SERPINB5
|
serpin family B member 5 |
| chr11_+_5389377 | 0.17 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr11_-_72793636 | 0.17 |
ENST00000538536.5
ENST00000543304.5 ENST00000540587.1 |
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr12_-_52926459 | 0.17 |
ENST00000552150.5
|
KRT8
|
keratin 8 |
| chr12_+_41437680 | 0.17 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr2_+_102311502 | 0.17 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr1_+_196774813 | 0.17 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr11_+_55811367 | 0.17 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr6_-_47042260 | 0.17 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chrX_+_66164340 | 0.17 |
ENST00000441993.7
ENST00000419594.6 ENST00000425114.2 |
HEPH
|
hephaestin |
| chr14_-_60169843 | 0.17 |
ENST00000536410.6
ENST00000216500.9 |
DHRS7
|
dehydrogenase/reductase 7 |
| chr14_+_22226711 | 0.16 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr12_+_56338873 | 0.16 |
ENST00000228534.6
|
IL23A
|
interleukin 23 subunit alpha |
| chr19_-_43656616 | 0.16 |
ENST00000593447.5
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_-_95072936 | 0.16 |
ENST00000370205.6
|
ALG14
|
ALG14 UDP-N-acetylglucosaminyltransferase subunit |
| chr6_+_111259294 | 0.16 |
ENST00000672303.1
ENST00000671876.2 ENST00000368847.5 |
MFSD4B
|
major facilitator superfamily domain containing 4B |
| chr3_+_122055355 | 0.16 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr6_-_28587250 | 0.16 |
ENST00000452236.3
|
ZBED9
|
zinc finger BED-type containing 9 |
| chr17_-_66229380 | 0.16 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr21_-_41926680 | 0.16 |
ENST00000329623.11
|
C2CD2
|
C2 calcium dependent domain containing 2 |
| chr1_+_152985231 | 0.16 |
ENST00000368762.1
|
SPRR1A
|
small proline rich protein 1A |
| chr1_+_240014319 | 0.16 |
ENST00000447095.5
|
FMN2
|
formin 2 |
| chrX_-_31178220 | 0.16 |
ENST00000681026.1
|
DMD
|
dystrophin |
| chr10_+_128047559 | 0.16 |
ENST00000306042.9
|
PTPRE
|
protein tyrosine phosphatase receptor type E |
| chr3_+_58008350 | 0.16 |
ENST00000490882.5
ENST00000358537.7 ENST00000429972.6 ENST00000682871.1 ENST00000295956.9 |
FLNB
|
filamin B |
| chr1_+_45688165 | 0.16 |
ENST00000372025.5
|
TMEM69
|
transmembrane protein 69 |
| chr19_+_41797147 | 0.15 |
ENST00000596544.1
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr9_-_101384999 | 0.15 |
ENST00000259407.7
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase |
| chr6_+_24356903 | 0.15 |
ENST00000274766.2
|
KAAG1
|
kidney associated antigen 1 |
| chr3_-_98522869 | 0.15 |
ENST00000502288.5
ENST00000512147.5 ENST00000341181.11 ENST00000510541.5 ENST00000503621.5 ENST00000511081.5 ENST00000507874.5 ENST00000502299.5 ENST00000508659.5 ENST00000510545.5 ENST00000511667.5 ENST00000394185.6 |
CLDND1
|
claudin domain containing 1 |
| chr7_-_32299287 | 0.15 |
ENST00000396193.5
|
PDE1C
|
phosphodiesterase 1C |
| chr11_-_72794032 | 0.15 |
ENST00000334805.11
|
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr3_+_112086364 | 0.15 |
ENST00000264848.10
|
C3orf52
|
chromosome 3 open reading frame 52 |
| chr11_-_124445696 | 0.15 |
ENST00000642064.1
|
OR8B8
|
olfactory receptor family 8 subfamily B member 8 |
| chr9_-_115091018 | 0.15 |
ENST00000542877.5
ENST00000537320.5 ENST00000341037.8 |
TNC
|
tenascin C |
| chr3_-_74521140 | 0.15 |
ENST00000263665.6
|
CNTN3
|
contactin 3 |
| chr3_-_169147734 | 0.15 |
ENST00000464456.5
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_+_99970430 | 0.15 |
ENST00000370153.6
|
SLC35A3
|
solute carrier family 35 member A3 |
| chr6_-_30744537 | 0.15 |
ENST00000259874.6
ENST00000376377.2 |
IER3
|
immediate early response 3 |
| chr4_+_88378733 | 0.15 |
ENST00000273960.7
ENST00000380265.9 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr3_-_197226351 | 0.15 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_218710821 | 0.15 |
ENST00000392102.6
ENST00000457313.5 ENST00000415717.5 |
TTLL4
|
tubulin tyrosine ligase like 4 |
| chr1_+_99850485 | 0.15 |
ENST00000370165.7
ENST00000370163.7 ENST00000294724.8 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr12_+_1970772 | 0.15 |
ENST00000682544.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr11_+_55635113 | 0.15 |
ENST00000641760.1
|
OR4P4
|
olfactory receptor family 4 subfamily P member 4 |
| chr14_+_21042352 | 0.15 |
ENST00000298690.5
|
RNASE7
|
ribonuclease A family member 7 |
| chr1_-_99766620 | 0.14 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr12_+_1970809 | 0.14 |
ENST00000683781.1
ENST00000682462.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr7_-_38249572 | 0.14 |
ENST00000436911.6
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr11_+_112176364 | 0.14 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr3_-_190322434 | 0.14 |
ENST00000295522.4
|
CLDN1
|
claudin 1 |
| chr7_+_116953482 | 0.14 |
ENST00000323984.8
ENST00000417919.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr11_-_7796942 | 0.14 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2 |
| chr1_-_10964201 | 0.14 |
ENST00000418570.6
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr1_-_153041111 | 0.14 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr18_+_58149314 | 0.14 |
ENST00000435432.6
ENST00000357895.9 ENST00000586263.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_+_160739286 | 0.14 |
ENST00000359331.8
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr12_-_10998304 | 0.14 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20 |
| chr18_+_58362467 | 0.14 |
ENST00000675101.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr7_+_116953514 | 0.14 |
ENST00000446490.5
|
ST7
|
suppression of tumorigenicity 7 |
| chr21_+_42199686 | 0.14 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr14_+_96797304 | 0.14 |
ENST00000553683.2
ENST00000680538.1 ENST00000681195.1 ENST00000679727.1 ENST00000680509.1 ENST00000557222.6 ENST00000680683.1 ENST00000680335.1 ENST00000216639.8 ENST00000679736.1 ENST00000681176.1 ENST00000557352.2 ENST00000679903.1 ENST00000681419.1 ENST00000680849.1 ENST00000681493.1 ENST00000679770.1 ENST00000681355.1 ENST00000681344.1 |
VRK1
|
VRK serine/threonine kinase 1 |
| chr1_+_15341744 | 0.14 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr9_-_136245802 | 0.14 |
ENST00000358701.10
|
QSOX2
|
quiescin sulfhydryl oxidase 2 |
| chr19_+_57830288 | 0.14 |
ENST00000442832.8
ENST00000594901.2 |
ZNF587B
|
zinc finger protein 587B |
| chr4_+_88378842 | 0.14 |
ENST00000264346.12
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr13_+_42781578 | 0.14 |
ENST00000313851.3
|
FAM216B
|
family with sequence similarity 216 member B |
| chr16_-_3372666 | 0.14 |
ENST00000399974.5
|
MTRNR2L4
|
MT-RNR2 like 4 |
| chr4_+_70334963 | 0.14 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr5_+_136058849 | 0.14 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr14_+_64715677 | 0.14 |
ENST00000634379.2
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chrX_+_17375230 | 0.14 |
ENST00000380060.7
|
NHS
|
NHS actin remodeling regulator |
| chrX_+_17375194 | 0.14 |
ENST00000676302.1
|
NHS
|
NHS actin remodeling regulator |
| chr7_+_116953238 | 0.14 |
ENST00000393446.6
|
ST7
|
suppression of tumorigenicity 7 |
| chr18_+_58196736 | 0.14 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.4 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.4 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.5 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.3 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.7 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.3 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.1 | 0.3 | GO:0002445 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.1 | 0.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.4 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.2 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.1 | 0.2 | GO:0043016 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.2 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) cellular response to lead ion(GO:0071284) positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.0 | 1.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.2 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.0 | 0.2 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.4 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.1 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.2 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.0 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0036404 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0002580 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.5 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:1902164 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 3.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0035739 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.0 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.0 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.0 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of T cell migration(GO:2000405) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.0 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 0.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.0 | GO:0060168 | regulation of adenosine receptor signaling pathway(GO:0060167) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.1 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.0 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.0 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.0 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.0 | GO:0035498 | carnosine metabolic process(GO:0035498) |
| 0.0 | 0.0 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0071753 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.0 | 0.1 | GO:0033167 | ARC complex(GO:0033167) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.0 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.2 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.0 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.0 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.0 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.0 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 2.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |