Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
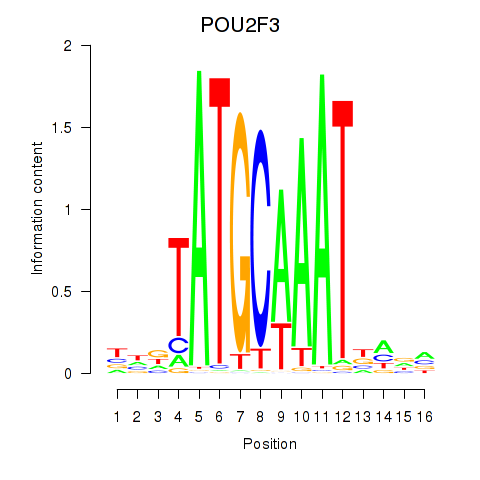
Results for POU5F1_POU2F3
Z-value: 0.75
Transcription factors associated with POU5F1_POU2F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
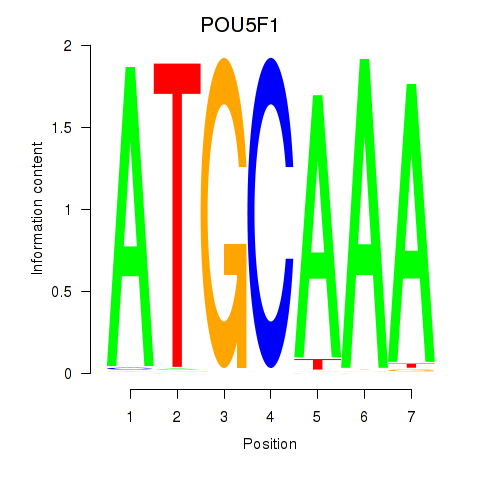
POU5F1
|
ENSG00000204531.20 | POU class 5 homeobox 1 |
|
POU2F3
|
ENSG00000137709.10 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU5F1 | hg38_v1_chr6_-_31170620_31170698 | -0.07 | 8.7e-01 | Click! |
| POU2F3 | hg38_v1_chr11_+_120240135_120240199, hg38_v1_chr11_+_120236635_120236642 | -0.05 | 9.0e-01 | Click! |
Activity profile of POU5F1_POU2F3 motif
Sorted Z-values of POU5F1_POU2F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_91946989 | 1.90 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr14_-_91947383 | 1.66 |
ENST00000267620.14
|
FBLN5
|
fibulin 5 |
| chr12_-_91180365 | 1.65 |
ENST00000547937.5
|
DCN
|
decorin |
| chr2_+_188974364 | 1.31 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr19_-_43619591 | 1.21 |
ENST00000598676.1
ENST00000300811.8 |
ZNF428
|
zinc finger protein 428 |
| chr12_-_91179355 | 1.08 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chrX_+_81202066 | 1.02 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chr5_+_120531464 | 1.00 |
ENST00000505123.5
|
PRR16
|
proline rich 16 |
| chr2_-_187554351 | 0.99 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr4_+_123399488 | 0.99 |
ENST00000394339.2
|
SPRY1
|
sprouty RTK signaling antagonist 1 |
| chr4_+_70050431 | 0.98 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
| chr14_-_91947654 | 0.95 |
ENST00000342058.9
|
FBLN5
|
fibulin 5 |
| chr5_-_111756245 | 0.90 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr2_+_240998608 | 0.88 |
ENST00000310397.13
|
SNED1
|
sushi, nidogen and EGF like domains 1 |
| chrX_-_107775951 | 0.84 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr12_-_91182652 | 0.79 |
ENST00000552145.5
ENST00000546745.5 |
DCN
|
decorin |
| chr12_-_91182784 | 0.77 |
ENST00000547568.6
ENST00000052754.10 ENST00000552962.5 |
DCN
|
decorin |
| chrX_-_107717054 | 0.77 |
ENST00000503515.1
ENST00000372397.6 |
TSC22D3
|
TSC22 domain family member 3 |
| chr19_-_49640092 | 0.76 |
ENST00000246792.4
|
RRAS
|
RAS related |
| chr20_+_34704336 | 0.72 |
ENST00000374809.6
ENST00000374810.8 ENST00000451665.5 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr12_-_91179472 | 0.63 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr12_+_6943811 | 0.62 |
ENST00000544681.1
ENST00000537087.5 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr6_-_35688907 | 0.62 |
ENST00000539068.5
ENST00000357266.9 |
FKBP5
|
FKBP prolyl isomerase 5 |
| chr12_+_6944009 | 0.60 |
ENST00000229281.6
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr9_-_92424427 | 0.60 |
ENST00000375550.5
|
OMD
|
osteomodulin |
| chr4_-_156970903 | 0.59 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr5_-_111757175 | 0.58 |
ENST00000509025.5
ENST00000257435.12 ENST00000515855.5 |
NREP
|
neuronal regeneration related protein |
| chr16_+_32995048 | 0.54 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chrX_-_81201886 | 0.53 |
ENST00000451455.1
ENST00000358130.7 ENST00000436386.5 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr12_+_6944065 | 0.53 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr1_-_145910066 | 0.53 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10 |
| chr1_-_145910031 | 0.52 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10 |
| chr1_+_163069353 | 0.52 |
ENST00000531057.5
ENST00000527809.5 ENST00000367908.8 ENST00000367909.11 |
RGS4
|
regulator of G protein signaling 4 |
| chr3_+_12351470 | 0.51 |
ENST00000287820.10
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr5_-_159099909 | 0.50 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1 |
| chr2_+_200308943 | 0.50 |
ENST00000619961.4
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr2_-_68319887 | 0.48 |
ENST00000409862.1
ENST00000263655.4 |
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr17_+_40062810 | 0.48 |
ENST00000584985.5
ENST00000264637.8 |
THRA
|
thyroid hormone receptor alpha |
| chr4_+_125314918 | 0.48 |
ENST00000674496.2
ENST00000394329.9 |
FAT4
|
FAT atypical cadherin 4 |
| chr10_-_77090722 | 0.46 |
ENST00000638531.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_+_61081728 | 0.46 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr3_+_12351493 | 0.44 |
ENST00000683699.1
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr21_-_38121331 | 0.43 |
ENST00000482032.1
ENST00000398948.5 ENST00000328264.7 ENST00000645093.1 |
DSCR4
KCNJ6
|
Down syndrome critical region 4 potassium inwardly rectifying channel subfamily J member 6 |
| chr1_+_61077219 | 0.42 |
ENST00000407417.7
|
NFIA
|
nuclear factor I A |
| chr2_-_189179754 | 0.41 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr7_-_13989658 | 0.40 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chr2_+_151357583 | 0.39 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr13_+_75804221 | 0.39 |
ENST00000489941.6
ENST00000525373.5 |
LMO7
|
LIM domain 7 |
| chr22_+_37696982 | 0.39 |
ENST00000644935.1
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr19_+_14583076 | 0.38 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr10_+_31319125 | 0.38 |
ENST00000320985.14
ENST00000560721.6 ENST00000558440.5 ENST00000424869.6 ENST00000542815.7 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr5_+_93583212 | 0.37 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr5_-_172771187 | 0.37 |
ENST00000239223.4
|
DUSP1
|
dual specificity phosphatase 1 |
| chr14_-_106130061 | 0.36 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr1_+_59310071 | 0.36 |
ENST00000371212.5
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr5_-_111976925 | 0.36 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr3_-_48556785 | 0.36 |
ENST00000232375.8
ENST00000383734.6 ENST00000416568.5 ENST00000412035.5 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr9_-_76906090 | 0.35 |
ENST00000376718.8
|
PRUNE2
|
prune homolog 2 with BCH domain |
| chr10_+_123148128 | 0.35 |
ENST00000339992.4
|
HMX2
|
H6 family homeobox 2 |
| chr20_+_41028814 | 0.35 |
ENST00000361337.3
|
TOP1
|
DNA topoisomerase I |
| chr7_-_13988863 | 0.35 |
ENST00000405358.8
|
ETV1
|
ETS variant transcription factor 1 |
| chr1_+_164559739 | 0.34 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1 |
| chr5_+_93584916 | 0.34 |
ENST00000647447.1
ENST00000615873.1 |
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr13_+_75804169 | 0.33 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr12_-_95548213 | 0.33 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr19_-_893172 | 0.33 |
ENST00000325464.6
ENST00000312090.10 |
MED16
|
mediator complex subunit 16 |
| chr2_-_178478499 | 0.32 |
ENST00000434643.6
|
FKBP7
|
FKBP prolyl isomerase 7 |
| chr9_-_76906041 | 0.32 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr2_-_187554473 | 0.32 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr17_-_68955332 | 0.32 |
ENST00000269080.6
ENST00000615593.4 ENST00000586539.6 ENST00000430352.6 |
ABCA8
|
ATP binding cassette subfamily A member 8 |
| chr1_-_243163310 | 0.31 |
ENST00000492145.1
ENST00000490813.5 ENST00000464936.5 |
CEP170
|
centrosomal protein 170 |
| chr22_-_28306645 | 0.31 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr19_-_18281612 | 0.31 |
ENST00000252818.5
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr19_-_893200 | 0.30 |
ENST00000269814.8
ENST00000395808.7 |
MED16
|
mediator complex subunit 16 |
| chr5_-_147453888 | 0.30 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr1_+_103655760 | 0.29 |
ENST00000370083.9
|
AMY1A
|
amylase alpha 1A |
| chr6_-_33711684 | 0.29 |
ENST00000374231.8
ENST00000607484.6 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr20_+_36214373 | 0.29 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr11_-_133845495 | 0.28 |
ENST00000299140.8
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr2_-_178478541 | 0.28 |
ENST00000424785.7
|
FKBP7
|
FKBP prolyl isomerase 7 |
| chr1_+_61082398 | 0.28 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A |
| chr4_-_185812209 | 0.28 |
ENST00000393523.6
ENST00000393528.7 ENST00000449407.6 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_17226662 | 0.28 |
ENST00000598068.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr5_-_173328407 | 0.28 |
ENST00000265087.9
|
STC2
|
stanniocalcin 2 |
| chr11_+_120240135 | 0.28 |
ENST00000543440.7
|
POU2F3
|
POU class 2 homeobox 3 |
| chr6_-_31684040 | 0.28 |
ENST00000375863.7
|
LY6G5C
|
lymphocyte antigen 6 family member G5C |
| chr14_-_106088573 | 0.28 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr9_-_120477354 | 0.27 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr17_-_7404039 | 0.27 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr2_-_174847015 | 0.27 |
ENST00000650938.1
|
CHN1
|
chimerin 1 |
| chr14_+_79279403 | 0.27 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr6_+_71886703 | 0.27 |
ENST00000491071.6
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_174846405 | 0.26 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr2_-_144517663 | 0.26 |
ENST00000427902.5
ENST00000462355.2 ENST00000470879.5 ENST00000409487.7 ENST00000435831.5 ENST00000630572.2 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_+_69492793 | 0.26 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr1_+_164559173 | 0.26 |
ENST00000420696.7
|
PBX1
|
PBX homeobox 1 |
| chr16_-_30096170 | 0.26 |
ENST00000566134.5
ENST00000565110.5 ENST00000398841.6 ENST00000398838.8 |
YPEL3
|
yippee like 3 |
| chr19_+_17226597 | 0.26 |
ENST00000597836.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr8_-_107497909 | 0.26 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1 |
| chr17_+_45135640 | 0.26 |
ENST00000586346.5
ENST00000321854.13 ENST00000398322.7 ENST00000592162.5 ENST00000376955.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr11_+_64318091 | 0.26 |
ENST00000265462.9
ENST00000352435.8 ENST00000347941.4 |
PRDX5
|
peroxiredoxin 5 |
| chr6_+_71886900 | 0.25 |
ENST00000517960.5
ENST00000518273.5 ENST00000522291.5 ENST00000521978.5 ENST00000520567.5 ENST00000264839.11 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr6_+_27147094 | 0.25 |
ENST00000377459.3
|
H2AC12
|
H2A clustered histone 12 |
| chr9_-_20382461 | 0.25 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr6_-_52995170 | 0.25 |
ENST00000370959.1
ENST00000370963.9 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr22_+_22704265 | 0.25 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr19_+_17406099 | 0.25 |
ENST00000634942.2
ENST00000634568.1 ENST00000600514.5 |
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator multivesicular body subunit 12A |
| chr19_+_39412650 | 0.24 |
ENST00000425673.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr3_-_149576203 | 0.24 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr19_+_17226218 | 0.24 |
ENST00000601529.5
ENST00000215061.9 ENST00000600232.5 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr3_-_112610262 | 0.24 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr10_-_62816341 | 0.24 |
ENST00000242480.4
ENST00000637191.1 |
EGR2
|
early growth response 2 |
| chr17_+_57257002 | 0.24 |
ENST00000322684.7
ENST00000579590.5 |
MSI2
|
musashi RNA binding protein 2 |
| chr2_-_38076076 | 0.23 |
ENST00000614273.1
ENST00000610745.5 ENST00000490576.1 |
CYP1B1
|
cytochrome P450 family 1 subfamily B member 1 |
| chr2_+_56183973 | 0.23 |
ENST00000407595.3
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr2_+_112911159 | 0.23 |
ENST00000263326.8
|
IL37
|
interleukin 37 |
| chr11_-_58844484 | 0.23 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr2_-_79086847 | 0.23 |
ENST00000454188.5
|
REG1B
|
regenerating family member 1 beta |
| chr1_+_32200587 | 0.23 |
ENST00000373602.10
ENST00000421922.6 ENST00000681230.1 |
CCDC28B
|
coiled-coil domain containing 28B |
| chr14_+_79279339 | 0.23 |
ENST00000557594.5
|
NRXN3
|
neurexin 3 |
| chr1_-_145885826 | 0.22 |
ENST00000544626.2
ENST00000355594.9 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr15_-_63156774 | 0.22 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like |
| chr1_-_160282458 | 0.22 |
ENST00000485079.1
|
ENSG00000258465.8
|
novel protein |
| chr1_+_6555301 | 0.22 |
ENST00000333172.11
ENST00000351136.7 |
TAS1R1
|
taste 1 receptor member 1 |
| chr3_+_148730100 | 0.22 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr14_+_79279681 | 0.22 |
ENST00000679122.1
|
NRXN3
|
neurexin 3 |
| chr12_+_59664677 | 0.22 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr4_-_158173042 | 0.22 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr10_+_94089067 | 0.22 |
ENST00000371375.1
ENST00000675218.1 |
PLCE1
|
phospholipase C epsilon 1 |
| chr5_+_61332236 | 0.22 |
ENST00000252744.6
|
ZSWIM6
|
zinc finger SWIM-type containing 6 |
| chr2_+_209579598 | 0.22 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr5_+_173918186 | 0.21 |
ENST00000657000.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr7_+_90709530 | 0.21 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr16_+_21678514 | 0.21 |
ENST00000286149.8
ENST00000388958.8 |
OTOA
|
otoancorin |
| chr10_-_62816309 | 0.21 |
ENST00000411732.3
|
EGR2
|
early growth response 2 |
| chr3_+_54123452 | 0.21 |
ENST00000620722.4
ENST00000490478.5 |
CACNA2D3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3 |
| chr1_+_65264694 | 0.21 |
ENST00000263441.11
ENST00000395325.7 |
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr15_-_19988117 | 0.21 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr20_-_62407274 | 0.21 |
ENST00000279101.9
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2 |
| chr1_+_47333863 | 0.21 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine/uridine monophosphate kinase 1 |
| chr1_+_164559766 | 0.21 |
ENST00000367897.5
ENST00000559240.5 |
PBX1
|
PBX homeobox 1 |
| chr6_-_33711717 | 0.21 |
ENST00000374214.3
|
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr5_+_173918216 | 0.21 |
ENST00000519467.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr19_+_35358460 | 0.21 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3 |
| chr15_-_58065734 | 0.20 |
ENST00000347587.7
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr15_-_58065870 | 0.20 |
ENST00000537372.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr2_+_219279330 | 0.20 |
ENST00000425450.5
ENST00000392086.8 ENST00000421532.5 ENST00000336576.10 |
DNAJB2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr5_-_39425187 | 0.20 |
ENST00000545653.5
|
DAB2
|
DAB adaptor protein 2 |
| chr6_-_15586006 | 0.20 |
ENST00000462989.6
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr12_-_7092422 | 0.20 |
ENST00000543835.5
ENST00000647956.2 ENST00000535233.6 |
C1R
|
complement C1r |
| chr2_-_40512361 | 0.20 |
ENST00000403092.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr6_-_24877262 | 0.20 |
ENST00000378023.8
ENST00000540914.5 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr22_+_22758698 | 0.20 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr15_-_58065703 | 0.20 |
ENST00000249750.9
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr19_+_3572971 | 0.20 |
ENST00000453933.5
ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr7_-_151248668 | 0.20 |
ENST00000262188.13
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr21_-_10649835 | 0.20 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr19_+_3572777 | 0.20 |
ENST00000416526.5
|
HMG20B
|
high mobility group 20B |
| chr19_-_12365628 | 0.20 |
ENST00000438182.5
|
ZNF442
|
zinc finger protein 442 |
| chr6_-_26033609 | 0.20 |
ENST00000615868.2
|
H2AC4
|
H2A clustered histone 4 |
| chr4_+_122339221 | 0.19 |
ENST00000442707.1
|
KIAA1109
|
KIAA1109 |
| chr1_+_103749898 | 0.19 |
ENST00000622339.5
|
AMY1C
|
amylase alpha 1C |
| chr13_-_42992165 | 0.19 |
ENST00000398762.7
ENST00000313640.11 ENST00000313624.12 |
EPSTI1
|
epithelial stromal interaction 1 |
| chr4_+_133149307 | 0.19 |
ENST00000618019.1
|
PCDH10
|
protocadherin 10 |
| chr2_+_161231078 | 0.19 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr14_-_106593319 | 0.19 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr7_+_90709231 | 0.19 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr3_+_141387616 | 0.19 |
ENST00000509883.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr12_+_133130618 | 0.19 |
ENST00000426665.6
ENST00000248211.11 |
ZNF10
|
zinc finger protein 10 |
| chr3_-_187745460 | 0.19 |
ENST00000406870.7
|
BCL6
|
BCL6 transcription repressor |
| chr10_+_81875173 | 0.19 |
ENST00000372141.7
ENST00000404547.5 |
NRG3
|
neuregulin 3 |
| chr19_+_926001 | 0.19 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr14_-_106811131 | 0.18 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr1_+_192809031 | 0.18 |
ENST00000235382.7
|
RGS2
|
regulator of G protein signaling 2 |
| chr12_-_10420550 | 0.18 |
ENST00000381903.2
ENST00000396439.7 |
KLRC3
|
killer cell lectin like receptor C3 |
| chr14_-_106675544 | 0.18 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chrX_-_136780925 | 0.18 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr22_-_23754376 | 0.18 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr6_+_101181254 | 0.18 |
ENST00000682090.1
ENST00000421544.6 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr22_+_43923755 | 0.18 |
ENST00000423180.2
ENST00000216180.8 |
PNPLA3
|
patatin like phospholipase domain containing 3 |
| chr11_+_20022550 | 0.18 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr9_+_18474206 | 0.18 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS like 1 |
| chrX_+_12975083 | 0.18 |
ENST00000451311.7
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4 X-linked |
| chr22_+_22818994 | 0.18 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 |
| chr22_-_19447686 | 0.18 |
ENST00000399568.5
ENST00000399562.9 |
C22orf39
|
chromosome 22 open reading frame 39 |
| chr2_-_151261839 | 0.18 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43 |
| chr3_-_15797930 | 0.18 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr14_-_106658251 | 0.17 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr10_+_94089034 | 0.17 |
ENST00000676102.1
ENST00000371385.8 |
PLCE1
|
phospholipase C epsilon 1 |
| chr2_-_210171402 | 0.17 |
ENST00000281772.14
|
KANSL1L
|
KAT8 regulatory NSL complex subunit 1 like |
| chr3_-_127822455 | 0.17 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr15_+_67166019 | 0.17 |
ENST00000537194.6
|
SMAD3
|
SMAD family member 3 |
| chr5_-_160400025 | 0.17 |
ENST00000523213.1
ENST00000408953.4 |
ZBED8
|
zinc finger BED-type containing 8 |
| chr7_+_130293134 | 0.17 |
ENST00000445470.6
ENST00000492072.5 ENST00000222482.10 ENST00000473956.5 ENST00000493259.5 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr1_-_3650063 | 0.17 |
ENST00000419924.2
ENST00000270708.12 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr5_+_172834225 | 0.17 |
ENST00000393784.8
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr1_+_248445512 | 0.17 |
ENST00000642130.1
ENST00000641925.2 |
OR2T2
|
olfactory receptor family 2 subfamily T member 2 |
| chr10_+_80132591 | 0.17 |
ENST00000372267.6
|
PLAC9
|
placenta associated 9 |
| chr1_-_167090370 | 0.17 |
ENST00000367868.4
|
GPA33
|
glycoprotein A33 |
| chr9_+_18474100 | 0.17 |
ENST00000327883.11
ENST00000431052.6 ENST00000380570.8 ENST00000380548.9 |
ADAMTSL1
|
ADAMTS like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU5F1_POU2F3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 4.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.0 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 1.7 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.2 | 1.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.5 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 1.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 1.0 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.4 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.1 | 0.2 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.2 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 1.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0070476 | RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.2 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) sclerotome development(GO:0061056) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.1 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:2000449 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) regulation of T cell extravasation(GO:2000407) positive regulation of monocyte extravasation(GO:2000439) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.2 | GO:0048294 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.4 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:2000364 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0032712 | regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of interleukin-3 production(GO:0032712) negative regulation of granulocyte colony-stimulating factor production(GO:0071656) negative regulation of macrophage colony-stimulating factor production(GO:1901257) |
| 0.0 | 0.1 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.5 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0070676 | regulation of multivesicular body size(GO:0010796) intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) uracil metabolic process(GO:0019860) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 1.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0061011 | gall bladder development(GO:0061010) hepatic duct development(GO:0061011) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.0 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:1900212 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0044007 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:0061084 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 5.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.6 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 1.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.1 | 4.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.2 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 1.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0036042 | lysophosphatidic acid binding(GO:0035727) long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.0 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 5.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.3 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 2.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |