|
chr18_+_23949847
Show fit
|
1.40 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3
|
|
chr12_-_8662619
Show fit
|
1.37 |
ENST00000544889.1
ENST00000543369.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr12_-_8662703
Show fit
|
1.33 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr6_-_136526177
Show fit
|
1.02 |
ENST00000617204.4
|
MAP7
|
microtubule associated protein 7
|
|
chr11_+_18266254
Show fit
|
1.00 |
ENST00000532858.5
ENST00000649195.1
ENST00000356524.9
ENST00000405158.2
|
SAA1
|
serum amyloid A1
|
|
chr6_-_136526472
Show fit
|
0.94 |
ENST00000454590.5
ENST00000432797.6
|
MAP7
|
microtubule associated protein 7
|
|
chr12_-_57767057
Show fit
|
0.87 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1
|
|
chr6_-_136525961
Show fit
|
0.85 |
ENST00000438100.6
|
MAP7
|
microtubule associated protein 7
|
|
chr12_-_8662808
Show fit
|
0.71 |
ENST00000359478.7
ENST00000396549.6
|
MFAP5
|
microfibril associated protein 5
|
|
chr1_-_186680411
Show fit
|
0.67 |
ENST00000367468.10
|
PTGS2
|
prostaglandin-endoperoxide synthase 2
|
|
chr18_+_36297661
Show fit
|
0.65 |
ENST00000257209.8
ENST00000590592.5
ENST00000359247.8
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr12_-_85836372
Show fit
|
0.65 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9
|
|
chr7_+_86643902
Show fit
|
0.61 |
ENST00000361669.7
|
GRM3
|
glutamate metabotropic receptor 3
|
|
chr1_+_24319511
Show fit
|
0.60 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3
|
|
chr2_+_162318884
Show fit
|
0.57 |
ENST00000446271.5
ENST00000429691.6
|
GCA
|
grancalcin
|
|
chr2_-_162318475
Show fit
|
0.53 |
ENST00000648433.1
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr1_+_24319342
Show fit
|
0.51 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3
|
|
chr19_-_4338786
Show fit
|
0.50 |
ENST00000601482.1
ENST00000600324.5
ENST00000594605.6
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr18_+_58149314
Show fit
|
0.50 |
ENST00000435432.6
ENST00000357895.9
ENST00000586263.5
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr15_-_78944985
Show fit
|
0.47 |
ENST00000615999.5
ENST00000677789.1
ENST00000676880.1
ENST00000677936.1
ENST00000220166.10
ENST00000677810.1
ENST00000678644.1
ENST00000677534.1
ENST00000677316.1
|
CTSH
|
cathepsin H
|
|
chr1_-_24187246
Show fit
|
0.47 |
ENST00000374421.7
ENST00000374418.3
ENST00000327575.6
ENST00000327535.6
|
IFNLR1
|
interferon lambda receptor 1
|
|
chr11_+_34621065
Show fit
|
0.46 |
ENST00000257831.8
|
EHF
|
ETS homologous factor
|
|
chr3_+_113947901
Show fit
|
0.46 |
ENST00000330212.7
ENST00000498275.5
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23
|
|
chr11_+_72114840
Show fit
|
0.44 |
ENST00000622388.4
|
FOLR3
|
folate receptor gamma
|
|
chr12_+_56079843
Show fit
|
0.43 |
ENST00000549282.5
ENST00000549061.5
ENST00000683059.1
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3
|
|
chr6_+_137867241
Show fit
|
0.43 |
ENST00000612899.5
ENST00000420009.5
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr6_-_11779606
Show fit
|
0.42 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein
|
|
chr12_+_82686889
Show fit
|
0.42 |
ENST00000321196.8
|
TMTC2
|
transmembrane O-mannosyltransferase targeting cadherins 2
|
|
chr12_-_8662881
Show fit
|
0.42 |
ENST00000433590.6
|
MFAP5
|
microfibril associated protein 5
|
|
chr6_+_137867414
Show fit
|
0.42 |
ENST00000237289.8
ENST00000433680.1
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr6_+_14117764
Show fit
|
0.42 |
ENST00000379153.4
|
CD83
|
CD83 molecule
|
|
chr6_+_116461364
Show fit
|
0.40 |
ENST00000368606.7
ENST00000368605.3
|
CALHM6
|
calcium homeostasis modulator family member 6
|
|
chr19_+_14031746
Show fit
|
0.40 |
ENST00000263379.4
|
IL27RA
|
interleukin 27 receptor subunit alpha
|
|
chr5_-_16508951
Show fit
|
0.39 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr5_-_16508858
Show fit
|
0.39 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr5_-_16508812
Show fit
|
0.38 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr3_+_113948004
Show fit
|
0.37 |
ENST00000638807.2
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23
|
|
chr12_-_12562851
Show fit
|
0.36 |
ENST00000298573.9
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr5_-_16508990
Show fit
|
0.35 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1
|
|
chr15_+_90184912
Show fit
|
0.35 |
ENST00000561085.1
ENST00000332496.10
|
SEMA4B
|
semaphorin 4B
|
|
chr5_-_16508788
Show fit
|
0.34 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr1_-_32901330
Show fit
|
0.34 |
ENST00000329151.5
ENST00000373463.8
|
TMEM54
|
transmembrane protein 54
|
|
chr1_-_112711355
Show fit
|
0.34 |
ENST00000606505.5
ENST00000605933.5
|
ENSG00000271810.5
|
novel protein, PPM1J-RHOC readthrough
|
|
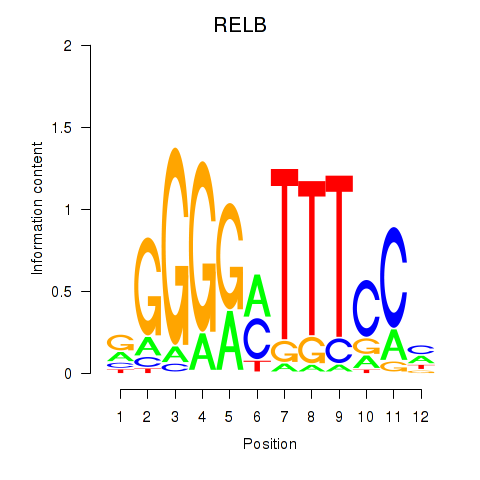
chr19_+_45001430
Show fit
|
0.33 |
ENST00000625761.2
ENST00000505236.1
ENST00000221452.13
|
RELB
|
RELB proto-oncogene, NF-kB subunit
|
|
chr1_-_183590876
Show fit
|
0.33 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr16_-_57284654
Show fit
|
0.32 |
ENST00000613167.4
ENST00000219207.10
ENST00000569059.5
|
PLLP
|
plasmolipin
|
|
chr11_+_34621109
Show fit
|
0.32 |
ENST00000450654.6
|
EHF
|
ETS homologous factor
|
|
chr8_+_100158576
Show fit
|
0.31 |
ENST00000388798.7
|
SPAG1
|
sperm associated antigen 1
|
|
chr11_+_102317492
Show fit
|
0.31 |
ENST00000673846.1
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr4_+_102501885
Show fit
|
0.30 |
ENST00000505458.5
|
NFKB1
|
nuclear factor kappa B subunit 1
|
|
chr2_+_112977998
Show fit
|
0.30 |
ENST00000259205.5
ENST00000376489.6
|
IL36G
|
interleukin 36 gamma
|
|
chr21_+_45455514
Show fit
|
0.30 |
ENST00000355480.10
ENST00000359759.8
|
COL18A1
|
collagen type XVIII alpha 1 chain
|
|
chr8_-_80171106
Show fit
|
0.29 |
ENST00000519303.6
|
TPD52
|
tumor protein D52
|
|
chr2_+_64454506
Show fit
|
0.28 |
ENST00000409537.2
|
LGALSL
|
galectin like
|
|
chr12_-_12562344
Show fit
|
0.27 |
ENST00000228862.3
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr8_-_65842051
Show fit
|
0.26 |
ENST00000401827.8
|
PDE7A
|
phosphodiesterase 7A
|
|
chr12_+_6946468
Show fit
|
0.25 |
ENST00000543115.5
ENST00000399448.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6
|
|
chr1_+_37474572
Show fit
|
0.25 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr10_-_71773513
Show fit
|
0.25 |
ENST00000394957.8
|
VSIR
|
V-set immunoregulatory receptor
|
|
chr2_+_158968608
Show fit
|
0.24 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chrX_-_24647091
Show fit
|
0.24 |
ENST00000356768.8
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr1_+_156153568
Show fit
|
0.24 |
ENST00000368284.5
ENST00000368286.6
ENST00000368285.8
ENST00000438830.5
|
SEMA4A
|
semaphorin 4A
|
|
chr1_-_6261053
Show fit
|
0.23 |
ENST00000377893.3
|
GPR153
|
G protein-coupled receptor 153
|
|
chr7_-_22500152
Show fit
|
0.22 |
ENST00000406890.6
ENST00000678116.1
ENST00000424363.5
|
STEAP1B
|
STEAP family member 1B
|
|
chr2_+_26346086
Show fit
|
0.22 |
ENST00000613142.4
ENST00000260585.12
ENST00000447170.1
|
SELENOI
|
selenoprotein I
|
|
chr6_+_29723340
Show fit
|
0.22 |
ENST00000334668.8
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr16_+_2009870
Show fit
|
0.22 |
ENST00000567649.1
|
NPW
|
neuropeptide W
|
|
chr2_-_88947820
Show fit
|
0.22 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5
|
|
chr1_+_45688165
Show fit
|
0.22 |
ENST00000372025.5
|
TMEM69
|
transmembrane protein 69
|
|
chr3_-_161104816
Show fit
|
0.22 |
ENST00000417187.2
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr2_+_60881515
Show fit
|
0.22 |
ENST00000295025.12
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr4_+_73869385
Show fit
|
0.21 |
ENST00000395761.4
|
CXCL1
|
C-X-C motif chemokine ligand 1
|
|
chr1_-_205775449
Show fit
|
0.21 |
ENST00000235932.8
ENST00000437324.6
ENST00000414729.1
ENST00000367139.8
|
RAB29
|
RAB29, member RAS oncogene family
|
|
chr2_-_162318613
Show fit
|
0.21 |
ENST00000649979.2
ENST00000421365.2
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr3_+_50205254
Show fit
|
0.21 |
ENST00000614032.5
ENST00000445096.5
|
SLC38A3
|
solute carrier family 38 member 3
|
|
chr8_+_22053543
Show fit
|
0.20 |
ENST00000519850.5
ENST00000381470.7
|
DMTN
|
dematin actin binding protein
|
|
chr19_+_40717091
Show fit
|
0.20 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr17_+_7380534
Show fit
|
0.20 |
ENST00000570896.5
|
TNK1
|
tyrosine kinase non receptor 1
|
|
chr11_-_124441158
Show fit
|
0.20 |
ENST00000328064.2
|
OR8B8
|
olfactory receptor family 8 subfamily B member 8
|
|
chr10_+_102394488
Show fit
|
0.20 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr3_-_52056552
Show fit
|
0.20 |
ENST00000495880.2
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr14_-_35404650
Show fit
|
0.20 |
ENST00000553342.1
ENST00000557140.5
ENST00000216797.10
|
NFKBIA
|
NFKB inhibitor alpha
|
|
chr2_+_60881553
Show fit
|
0.19 |
ENST00000394479.4
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chrX_-_20266606
Show fit
|
0.19 |
ENST00000643085.1
ENST00000645270.1
ENST00000644368.1
|
RPS6KA3
|
ribosomal protein S6 kinase A3
|
|
chr6_+_29723421
Show fit
|
0.19 |
ENST00000259951.12
ENST00000434407.6
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr1_+_156149657
Show fit
|
0.19 |
ENST00000414683.5
|
SEMA4A
|
semaphorin 4A
|
|
chr4_-_184474518
Show fit
|
0.19 |
ENST00000393593.8
|
IRF2
|
interferon regulatory factor 2
|
|
chr1_-_205775182
Show fit
|
0.19 |
ENST00000446390.6
|
RAB29
|
RAB29, member RAS oncogene family
|
|
chr6_-_10838467
Show fit
|
0.19 |
ENST00000313243.6
|
MAK
|
male germ cell associated kinase
|
|
chr6_-_24358036
Show fit
|
0.19 |
ENST00000378454.8
|
DCDC2
|
doublecortin domain containing 2
|
|
chr14_+_23372809
Show fit
|
0.18 |
ENST00000397242.2
ENST00000329715.2
|
IL25
|
interleukin 25
|
|
chr14_+_103123452
Show fit
|
0.18 |
ENST00000558056.1
ENST00000560869.6
|
TNFAIP2
|
TNF alpha induced protein 2
|
|
chr6_-_10838503
Show fit
|
0.18 |
ENST00000536370.6
ENST00000474039.5
ENST00000354489.7
ENST00000676116.1
|
MAK
|
male germ cell associated kinase
|
|
chr19_-_4831689
Show fit
|
0.18 |
ENST00000248244.6
|
TICAM1
|
toll like receptor adaptor molecule 1
|
|
chr10_+_132397168
Show fit
|
0.18 |
ENST00000631148.2
ENST00000305233.6
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr15_+_85380625
Show fit
|
0.18 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13
|
|
chr1_+_23791138
Show fit
|
0.17 |
ENST00000374514.8
ENST00000420982.5
ENST00000374505.6
|
LYPLA2
|
lysophospholipase 2
|
|
chr11_+_33376077
Show fit
|
0.17 |
ENST00000658780.2
|
KIAA1549L
|
KIAA1549 like
|
|
chr16_+_2019777
Show fit
|
0.17 |
ENST00000566435.4
|
NPW
|
neuropeptide W
|
|
chr7_-_111562455
Show fit
|
0.16 |
ENST00000452895.5
ENST00000405709.7
ENST00000452753.1
ENST00000331762.7
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2
|
|
chr4_-_73998669
Show fit
|
0.16 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5
|
|
chr6_+_29942523
Show fit
|
0.16 |
ENST00000376809.10
ENST00000376802.2
ENST00000638375.1
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr20_+_9514562
Show fit
|
0.16 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5
|
|
chr11_+_102317542
Show fit
|
0.15 |
ENST00000532808.5
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr15_-_30991415
Show fit
|
0.15 |
ENST00000563714.5
|
MTMR10
|
myotubularin related protein 10
|
|
chr4_+_77605807
Show fit
|
0.15 |
ENST00000682537.1
|
CXCL13
|
C-X-C motif chemokine ligand 13
|
|
chr14_+_91114431
Show fit
|
0.15 |
ENST00000428926.6
ENST00000517362.5
|
DGLUCY
|
D-glutamate cyclase
|
|
chr12_+_6199715
Show fit
|
0.15 |
ENST00000382518.6
ENST00000642746.1
ENST00000538834.6
|
CD9
|
CD9 molecule
|
|
chr16_+_50696999
Show fit
|
0.15 |
ENST00000300589.6
|
NOD2
|
nucleotide binding oligomerization domain containing 2
|
|
chr3_+_6861107
Show fit
|
0.15 |
ENST00000357716.9
ENST00000486284.5
ENST00000389336.8
|
GRM7
|
glutamate metabotropic receptor 7
|
|
chr1_-_1421302
Show fit
|
0.14 |
ENST00000520296.5
|
ANKRD65
|
ankyrin repeat domain 65
|
|
chr14_+_102777555
Show fit
|
0.14 |
ENST00000539721.5
ENST00000560463.5
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr21_-_41953997
Show fit
|
0.14 |
ENST00000380486.4
|
C2CD2
|
C2 calcium dependent domain containing 2
|
|
chr2_+_64453969
Show fit
|
0.14 |
ENST00000464281.5
|
LGALSL
|
galectin like
|
|
chr10_+_18659382
Show fit
|
0.14 |
ENST00000377275.4
|
ARL5B
|
ADP ribosylation factor like GTPase 5B
|
|
chr12_-_54981838
Show fit
|
0.13 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1
|
|
chr11_+_102317450
Show fit
|
0.13 |
ENST00000615299.4
ENST00000527309.2
ENST00000526421.6
ENST00000263464.9
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr17_-_76551174
Show fit
|
0.13 |
ENST00000589145.1
|
CYGB
|
cytoglobin
|
|
chr14_+_91114364
Show fit
|
0.13 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase
|
|
chrX_+_51406947
Show fit
|
0.13 |
ENST00000342995.4
|
EZHIP
|
EZH inhibitory protein
|
|
chr6_+_15248855
Show fit
|
0.13 |
ENST00000397311.4
|
JARID2
|
jumonji and AT-rich interaction domain containing 2
|
|
chr9_-_32526185
Show fit
|
0.13 |
ENST00000379883.3
ENST00000379868.6
ENST00000679859.1
|
DDX58
|
DExD/H-box helicase 58
|
|
chr15_+_34102037
Show fit
|
0.13 |
ENST00000397766.4
|
PGBD4
|
piggyBac transposable element derived 4
|
|
chr14_+_102777461
Show fit
|
0.13 |
ENST00000560371.5
ENST00000347662.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr16_-_2009797
Show fit
|
0.13 |
ENST00000563630.6
ENST00000562103.2
|
ZNF598
|
zinc finger protein 598, E3 ubiquitin ligase
|
|
chr12_+_2053545
Show fit
|
0.13 |
ENST00000682336.1
ENST00000344100.7
ENST00000399591.5
ENST00000399595.5
ENST00000399597.5
ENST00000399601.5
ENST00000399606.5
ENST00000399621.5
ENST00000399629.5
ENST00000399637.5
ENST00000399638.5
ENST00000399644.5
ENST00000399649.5
ENST00000402845.7
ENST00000682686.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C
|
|
chr17_-_48722510
Show fit
|
0.13 |
ENST00000290294.5
|
PRAC1
|
PRAC1 small nuclear protein
|
|
chr11_-_5154757
Show fit
|
0.13 |
ENST00000380367.3
|
OR52A1
|
olfactory receptor family 52 subfamily A member 1
|
|
chr8_-_33567118
Show fit
|
0.12 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122
|
|
chr17_-_6556447
Show fit
|
0.12 |
ENST00000421306.7
|
PITPNM3
|
PITPNM family member 3
|
|
chr2_+_165572329
Show fit
|
0.12 |
ENST00000342316.8
|
CSRNP3
|
cysteine and serine rich nuclear protein 3
|
|
chr20_-_4815100
Show fit
|
0.12 |
ENST00000379376.2
|
RASSF2
|
Ras association domain family member 2
|
|
chr22_-_40819324
Show fit
|
0.12 |
ENST00000435456.7
ENST00000434185.1
ENST00000544408.5
|
SLC25A17
|
solute carrier family 25 member 17
|
|
chr18_+_46946821
Show fit
|
0.12 |
ENST00000245121.9
ENST00000356157.12
|
KATNAL2
|
katanin catalytic subunit A1 like 2
|
|
chr1_-_204411804
Show fit
|
0.12 |
ENST00000367188.5
|
PPP1R15B
|
protein phosphatase 1 regulatory subunit 15B
|
|
chr12_+_100473916
Show fit
|
0.12 |
ENST00000549996.5
|
NR1H4
|
nuclear receptor subfamily 1 group H member 4
|
|
chr1_+_46247684
Show fit
|
0.12 |
ENST00000463715.5
ENST00000442598.5
ENST00000493985.5
ENST00000671528.1
|
RAD54L
|
RAD54 like
|
|
chr1_-_155990062
Show fit
|
0.12 |
ENST00000462460.6
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2
|
|
chr6_-_29559724
Show fit
|
0.11 |
ENST00000377050.5
|
UBD
|
ubiquitin D
|
|
chr1_+_202010575
Show fit
|
0.11 |
ENST00000367283.7
ENST00000367284.10
|
ELF3
|
E74 like ETS transcription factor 3
|
|
chr6_+_135181268
Show fit
|
0.11 |
ENST00000341911.10
ENST00000442647.7
ENST00000618728.4
ENST00000316528.12
ENST00000616088.4
|
MYB
|
MYB proto-oncogene, transcription factor
|
|
chr19_-_40716869
Show fit
|
0.11 |
ENST00000677018.1
ENST00000324464.8
ENST00000594720.6
ENST00000677496.1
|
COQ8B
|
coenzyme Q8B
|
|
chr6_+_5261497
Show fit
|
0.11 |
ENST00000274680.9
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial
|
|
chr1_+_156642108
Show fit
|
0.11 |
ENST00000457777.6
ENST00000329117.10
ENST00000424639.5
|
BCAN
|
brevican
|
|
chr8_-_17002327
Show fit
|
0.11 |
ENST00000180166.6
|
FGF20
|
fibroblast growth factor 20
|
|
chr5_-_88883701
Show fit
|
0.11 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_+_123845048
Show fit
|
0.10 |
ENST00000488323.5
ENST00000223026.9
|
HYAL4
|
hyaluronidase 4
|
|
chr16_-_74985070
Show fit
|
0.10 |
ENST00000616369.4
ENST00000262144.11
|
WDR59
|
WD repeat domain 59
|
|
chr16_+_30023198
Show fit
|
0.10 |
ENST00000681219.1
ENST00000300575.6
|
C16orf92
|
chromosome 16 open reading frame 92
|
|
chr14_+_102777433
Show fit
|
0.10 |
ENST00000392745.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr2_+_233693659
Show fit
|
0.10 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6
|
|
chr1_+_23791203
Show fit
|
0.10 |
ENST00000374503.7
ENST00000374502.7
|
LYPLA2
|
lysophospholipase 2
|
|
chr22_-_37984534
Show fit
|
0.10 |
ENST00000396884.8
|
SOX10
|
SRY-box transcription factor 10
|
|
chr1_+_46247731
Show fit
|
0.10 |
ENST00000371975.9
ENST00000469835.6
|
RAD54L
|
RAD54 like
|
|
chr16_-_66925526
Show fit
|
0.10 |
ENST00000299759.11
ENST00000420652.5
|
RRAD
|
RRAD, Ras related glycolysis inhibitor and calcium channel regulator
|
|
chr14_+_22829879
Show fit
|
0.10 |
ENST00000355151.9
ENST00000397496.7
ENST00000555345.5
ENST00000432849.7
ENST00000553711.5
ENST00000556465.5
ENST00000397505.2
ENST00000557221.1
ENST00000556840.5
ENST00000555536.1
|
MRPL52
|
mitochondrial ribosomal protein L52
|
|
chr10_+_122461545
Show fit
|
0.10 |
ENST00000368984.8
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr1_+_25338294
Show fit
|
0.10 |
ENST00000374358.5
|
TMEM50A
|
transmembrane protein 50A
|
|
chr15_-_40307910
Show fit
|
0.09 |
ENST00000543785.3
ENST00000260402.8
|
PLCB2
|
phospholipase C beta 2
|
|
chr12_-_121580954
Show fit
|
0.09 |
ENST00000536437.5
ENST00000611216.4
ENST00000538046.6
ENST00000377071.9
|
KDM2B
|
lysine demethylase 2B
|
|
chr12_-_124989053
Show fit
|
0.09 |
ENST00000308736.7
|
DHX37
|
DEAH-box helicase 37
|
|
chr12_+_103930332
Show fit
|
0.09 |
ENST00000681861.1
ENST00000550595.2
ENST00000680762.1
ENST00000614327.2
ENST00000681949.1
ENST00000299767.10
|
HSP90B1
|
heat shock protein 90 beta family member 1
|
|
chr5_+_141177790
Show fit
|
0.09 |
ENST00000239444.4
ENST00000623995.1
|
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8
novel transcript
|
|
chr1_+_53894181
Show fit
|
0.09 |
ENST00000361921.8
ENST00000322679.10
ENST00000613679.4
ENST00000617230.2
ENST00000610607.4
ENST00000532493.5
ENST00000525202.5
ENST00000524406.5
ENST00000388876.3
|
DIO1
|
iodothyronine deiodinase 1
|
|
chr1_+_161258738
Show fit
|
0.09 |
ENST00000504449.2
|
PCP4L1
|
Purkinje cell protein 4 like 1
|
|
chr4_-_46124046
Show fit
|
0.09 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1
|
|
chr1_-_153540694
Show fit
|
0.09 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5
|
|
chr20_-_34303345
Show fit
|
0.09 |
ENST00000217426.7
|
AHCY
|
adenosylhomocysteinase
|
|
chr12_-_117099434
Show fit
|
0.09 |
ENST00000541210.5
|
TESC
|
tescalcin
|
|
chr1_+_151047699
Show fit
|
0.09 |
ENST00000368926.6
|
C1orf56
|
chromosome 1 open reading frame 56
|
|
chr9_-_127755243
Show fit
|
0.09 |
ENST00000629203.2
ENST00000420366.5
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr12_-_117099384
Show fit
|
0.09 |
ENST00000335209.12
|
TESC
|
tescalcin
|
|
chr14_+_52267683
Show fit
|
0.09 |
ENST00000306051.3
ENST00000553372.1
|
PTGDR
|
prostaglandin D2 receptor
|
|
chr4_-_74099187
Show fit
|
0.09 |
ENST00000508487.3
|
CXCL2
|
C-X-C motif chemokine ligand 2
|
|
chr4_-_78939352
Show fit
|
0.09 |
ENST00000512733.5
|
PAQR3
|
progestin and adipoQ receptor family member 3
|
|
chr21_+_33403466
Show fit
|
0.09 |
ENST00000405436.5
|
IFNGR2
|
interferon gamma receptor 2
|
|
chr17_+_12665882
Show fit
|
0.08 |
ENST00000425538.6
|
MYOCD
|
myocardin
|
|
chr8_-_123274255
Show fit
|
0.08 |
ENST00000622816.2
ENST00000395571.8
|
ZHX1-C8orf76
ZHX1
|
ZHX1-C8orf76 readthrough
zinc fingers and homeoboxes 1
|
|
chr7_+_69599588
Show fit
|
0.08 |
ENST00000403018.3
|
AUTS2
|
activator of transcription and developmental regulator AUTS2
|
|
chr2_+_89913982
Show fit
|
0.08 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33
|
|
chr5_-_151080978
Show fit
|
0.08 |
ENST00000520931.5
ENST00000521591.6
ENST00000520695.5
ENST00000610535.5
ENST00000518977.5
ENST00000389378.6
ENST00000610874.4
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr5_+_141199555
Show fit
|
0.08 |
ENST00000624887.1
ENST00000354757.5
|
PCDHB11
|
protocadherin beta 11
|
|
chr1_+_42463221
Show fit
|
0.08 |
ENST00000654683.1
ENST00000667205.1
ENST00000655164.1
ENST00000657597.1
ENST00000667947.1
ENST00000668663.1
ENST00000660083.1
ENST00000655845.1
ENST00000671281.1
ENST00000664805.1
ENST00000654604.1
ENST00000655447.1
ENST00000661864.1
ENST00000665176.1
ENST00000670982.1
ENST00000668036.1
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr9_+_38392694
Show fit
|
0.08 |
ENST00000377698.4
ENST00000635162.1
|
ALDH1B1
|
aldehyde dehydrogenase 1 family member B1
|
|
chr12_+_2053311
Show fit
|
0.08 |
ENST00000399617.6
ENST00000683482.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C
|
|
chr17_-_79009731
Show fit
|
0.08 |
ENST00000392446.10
ENST00000590370.5
ENST00000591625.5
|
CANT1
|
calcium activated nucleotidase 1
|
|
chr6_+_138161932
Show fit
|
0.08 |
ENST00000251691.5
|
ARFGEF3
|
ARFGEF family member 3
|
|
chrX_+_48786578
Show fit
|
0.08 |
ENST00000376670.9
|
GATA1
|
GATA binding protein 1
|
|
chr16_-_70289480
Show fit
|
0.08 |
ENST00000261772.13
ENST00000675953.1
ENST00000675691.1
ENST00000675133.1
ENST00000674512.1
ENST00000675045.1
ENST00000675853.1
ENST00000565361.3
ENST00000674963.1
ENST00000674691.1
|
AARS1
|
alanyl-tRNA synthetase 1
|
|
chr2_-_100104530
Show fit
|
0.08 |
ENST00000432037.5
ENST00000673232.1
ENST00000423966.6
ENST00000409236.6
|
AFF3
|
AF4/FMR2 family member 3
|
|
chr17_-_40665121
Show fit
|
0.08 |
ENST00000394052.5
|
KRT222
|
keratin 222
|
|
chr5_+_151212117
Show fit
|
0.08 |
ENST00000523466.5
|
GM2A
|
GM2 ganglioside activator
|
|
chr7_-_102616692
Show fit
|
0.07 |
ENST00000521076.5
ENST00000462172.5
ENST00000522801.5
ENST00000262940.12
ENST00000449970.6
|
RASA4
|
RAS p21 protein activator 4
|
|
chrX_-_11665908
Show fit
|
0.07 |
ENST00000337414.9
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr14_+_91114026
Show fit
|
0.07 |
ENST00000521081.5
ENST00000520328.5
ENST00000524232.5
ENST00000522170.5
ENST00000256324.15
ENST00000519950.5
ENST00000523879.5
ENST00000521077.6
ENST00000518665.6
|
DGLUCY
|
D-glutamate cyclase
|
|
chr8_+_49911604
Show fit
|
0.07 |
ENST00000642164.1
ENST00000644093.1
ENST00000643999.1
ENST00000647073.1
ENST00000646880.1
|
SNTG1
|
syntrophin gamma 1
|
|
chr8_+_106726012
Show fit
|
0.07 |
ENST00000449762.6
ENST00000297447.10
|
OXR1
|
oxidation resistance 1
|
|
chr14_+_88824621
Show fit
|
0.07 |
ENST00000622513.4
ENST00000380656.7
ENST00000338104.10
ENST00000346301.8
ENST00000354441.10
ENST00000556651.5
ENST00000554686.5
|
TTC8
|
tetratricopeptide repeat domain 8
|
|
chr11_+_61248122
Show fit
|
0.07 |
ENST00000451616.6
|
PGA5
|
pepsinogen A5
|
|
chr5_+_127649018
Show fit
|
0.07 |
ENST00000379445.7
|
CTXN3
|
cortexin 3
|