Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for SHOX
Z-value: 0.99
Transcription factors associated with SHOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SHOX
|
ENSG00000185960.14 | short stature homeobox |
|
SHOX
|
ENSG00000185960.14 | short stature homeobox |
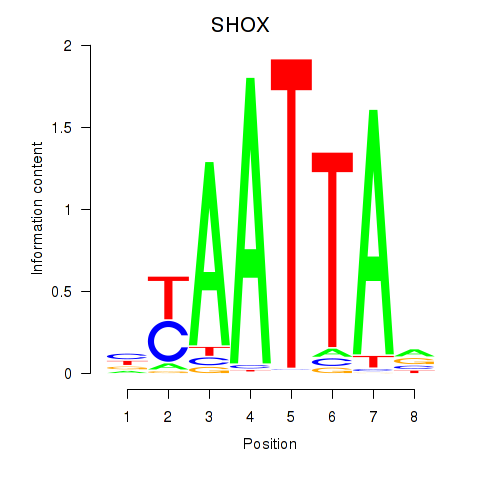
Activity profile of SHOX motif
Sorted Z-values of SHOX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_151316795 | 2.57 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr1_-_242449478 | 1.69 |
ENST00000427495.5
|
PLD5
|
phospholipase D family member 5 |
| chr19_-_51019699 | 1.42 |
ENST00000358789.8
|
KLK10
|
kallikrein related peptidase 10 |
| chr5_-_1882902 | 1.17 |
ENST00000231357.7
|
IRX4
|
iroquois homeobox 4 |
| chr7_-_41703062 | 1.17 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr12_-_27970273 | 1.16 |
ENST00000542963.1
ENST00000535992.5 |
PTHLH
|
parathyroid hormone like hormone |
| chr12_-_27970047 | 1.15 |
ENST00000395868.7
|
PTHLH
|
parathyroid hormone like hormone |
| chr19_-_51020154 | 1.09 |
ENST00000391805.5
ENST00000599077.1 |
KLK10
|
kallikrein related peptidase 10 |
| chr19_-_51020019 | 1.04 |
ENST00000309958.7
|
KLK10
|
kallikrein related peptidase 10 |
| chr8_+_32647080 | 1.03 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr15_+_100879822 | 1.00 |
ENST00000329841.10
ENST00000557963.1 ENST00000346623.6 |
ALDH1A3
|
aldehyde dehydrogenase 1 family member A3 |
| chr5_+_53480619 | 0.94 |
ENST00000396947.7
ENST00000256759.8 |
FST
|
follistatin |
| chr7_-_22193824 | 0.86 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr7_-_22194709 | 0.85 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr6_-_136466858 | 0.81 |
ENST00000544465.5
|
MAP7
|
microtubule associated protein 7 |
| chr18_+_36544544 | 0.78 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr13_-_85799400 | 0.77 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr7_+_70596078 | 0.73 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr2_+_68734773 | 0.72 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_+_160400543 | 0.72 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr6_-_136526472 | 0.71 |
ENST00000454590.5
ENST00000432797.6 |
MAP7
|
microtubule associated protein 7 |
| chr8_+_32646838 | 0.70 |
ENST00000651333.1
ENST00000652592.1 |
NRG1
|
neuregulin 1 |
| chr12_-_27972725 | 0.69 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr11_-_117876892 | 0.69 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117876719 | 0.68 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr5_+_67004618 | 0.68 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_+_68447453 | 0.67 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr11_-_117877463 | 0.66 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr8_+_104223320 | 0.64 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_+_111999189 | 0.62 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr20_-_51802433 | 0.60 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr13_-_75366973 | 0.59 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chr3_+_111998915 | 0.58 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr3_+_111998739 | 0.57 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr3_+_111999326 | 0.57 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr12_-_52434363 | 0.56 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr16_+_69105636 | 0.56 |
ENST00000569188.6
|
HAS3
|
hyaluronan synthase 3 |
| chr4_-_73620391 | 0.55 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr11_-_124320197 | 0.54 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr9_+_12693327 | 0.52 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr12_+_41437680 | 0.52 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr5_+_36606355 | 0.51 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr4_-_25863537 | 0.51 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr17_+_41226648 | 0.51 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr4_+_76435216 | 0.50 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chrX_+_106693838 | 0.48 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr11_-_120138104 | 0.47 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr10_+_24466487 | 0.46 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr12_-_89352487 | 0.46 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr17_+_41237998 | 0.46 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr6_+_121437378 | 0.46 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr7_-_22193728 | 0.46 |
ENST00000620335.4
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr17_-_41118369 | 0.45 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr8_-_17676484 | 0.45 |
ENST00000634613.1
ENST00000519066.5 |
MTUS1
|
microtubule associated scaffold protein 1 |
| chr12_+_26195313 | 0.45 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr10_-_49762276 | 0.44 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr13_+_77535669 | 0.44 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr10_+_24208774 | 0.43 |
ENST00000376456.8
ENST00000458595.5 ENST00000376452.7 ENST00000430453.6 |
KIAA1217
|
KIAA1217 |
| chr10_-_49762335 | 0.42 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr8_+_104223344 | 0.42 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr21_-_40847149 | 0.42 |
ENST00000400454.6
|
DSCAM
|
DS cell adhesion molecule |
| chr10_-_102120246 | 0.42 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1 |
| chr13_+_77535681 | 0.41 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535742 | 0.41 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr1_+_153774210 | 0.41 |
ENST00000271857.6
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr3_-_142029108 | 0.40 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr12_-_89352395 | 0.39 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_31326111 | 0.39 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr2_+_68734861 | 0.39 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr7_-_25228485 | 0.38 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr3_-_191282383 | 0.36 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr8_-_42377227 | 0.36 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr13_-_46142834 | 0.35 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr12_-_31326142 | 0.34 |
ENST00000337682.9
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr4_-_122621011 | 0.34 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr14_-_53956811 | 0.34 |
ENST00000559087.5
ENST00000245451.9 |
BMP4
|
bone morphogenetic protein 4 |
| chr18_-_55351977 | 0.34 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr8_-_42501224 | 0.33 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr3_-_27722699 | 0.33 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr14_+_22497657 | 0.33 |
ENST00000390496.1
|
TRAJ41
|
T cell receptor alpha joining 41 |
| chr4_-_73620629 | 0.32 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr18_-_55586092 | 0.32 |
ENST00000563888.6
ENST00000540999.5 ENST00000627685.2 |
TCF4
|
transcription factor 4 |
| chr18_-_55585773 | 0.31 |
ENST00000563824.5
ENST00000626425.2 ENST00000566514.5 ENST00000568673.5 ENST00000562847.5 ENST00000568147.5 |
TCF4
|
transcription factor 4 |
| chr6_-_39725335 | 0.30 |
ENST00000538893.5
|
KIF6
|
kinesin family member 6 |
| chr3_-_27722316 | 0.30 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr8_+_100158576 | 0.30 |
ENST00000388798.7
|
SPAG1
|
sperm associated antigen 1 |
| chr12_+_19205294 | 0.29 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr11_-_16356538 | 0.29 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr10_-_102120318 | 0.29 |
ENST00000673968.1
|
LDB1
|
LIM domain binding 1 |
| chr4_-_142305935 | 0.29 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr12_+_26195543 | 0.29 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr3_+_130850585 | 0.28 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr4_+_94974984 | 0.28 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr20_-_52105644 | 0.28 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr3_-_52056552 | 0.28 |
ENST00000495880.2
|
DUSP7
|
dual specificity phosphatase 7 |
| chr10_+_132397168 | 0.28 |
ENST00000631148.2
ENST00000305233.6 |
PWWP2B
|
PWWP domain containing 2B |
| chr4_-_142305826 | 0.27 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr20_-_51802509 | 0.27 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr18_+_23992773 | 0.27 |
ENST00000304621.10
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr9_+_470291 | 0.27 |
ENST00000382303.5
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr6_+_130018565 | 0.27 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr17_-_66229380 | 0.27 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr7_+_18496269 | 0.26 |
ENST00000432645.6
|
HDAC9
|
histone deacetylase 9 |
| chr11_-_129192291 | 0.26 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr17_-_9791586 | 0.26 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chr12_-_31325494 | 0.25 |
ENST00000543615.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr18_+_58341038 | 0.25 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr12_-_31326171 | 0.25 |
ENST00000542983.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr3_+_122055355 | 0.24 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr17_-_40799939 | 0.24 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chrX_+_135520616 | 0.24 |
ENST00000370752.4
ENST00000639893.2 |
INTS6L
|
integrator complex subunit 6 like |
| chrX_+_105948429 | 0.24 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr2_+_161416273 | 0.23 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr6_+_26365176 | 0.23 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr1_+_107139996 | 0.23 |
ENST00000370073.6
ENST00000370074.8 |
NTNG1
|
netrin G1 |
| chr6_+_26365215 | 0.23 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr13_-_35476682 | 0.23 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chrX_+_108045050 | 0.22 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_+_158968608 | 0.22 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr6_+_26402237 | 0.22 |
ENST00000476549.6
ENST00000450085.6 ENST00000425234.6 ENST00000427334.5 ENST00000506698.1 ENST00000289361.11 |
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr14_+_22508602 | 0.22 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr6_+_26365159 | 0.22 |
ENST00000532865.5
ENST00000396934.7 ENST00000508906.6 ENST00000530653.5 ENST00000527417.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr13_+_108629605 | 0.22 |
ENST00000457511.7
|
MYO16
|
myosin XVI |
| chr3_+_4680617 | 0.21 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr8_-_20183127 | 0.21 |
ENST00000276373.10
ENST00000519026.5 ENST00000440926.3 ENST00000437980.3 |
SLC18A1
|
solute carrier family 18 member A1 |
| chrX_+_108044967 | 0.21 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_-_207167220 | 0.21 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr8_-_20183090 | 0.20 |
ENST00000265808.11
ENST00000522513.5 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr7_+_130344810 | 0.20 |
ENST00000497503.5
ENST00000463587.5 ENST00000461828.5 ENST00000474905.6 ENST00000494311.1 ENST00000466363.6 |
CPA5
|
carboxypeptidase A5 |
| chr8_-_108787563 | 0.20 |
ENST00000297459.4
|
TMEM74
|
transmembrane protein 74 |
| chr3_-_79767987 | 0.20 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1 |
| chr2_-_49974182 | 0.20 |
ENST00000412315.5
ENST00000378262.7 |
NRXN1
|
neurexin 1 |
| chr9_+_121567057 | 0.20 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr17_-_79950828 | 0.20 |
ENST00000572862.5
ENST00000573782.5 ENST00000574427.1 ENST00000570373.5 ENST00000340848.11 ENST00000576768.5 |
TBC1D16
|
TBC1 domain family member 16 |
| chr14_+_61187544 | 0.20 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr6_-_111606260 | 0.20 |
ENST00000340026.10
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr2_+_218270392 | 0.19 |
ENST00000248451.7
ENST00000273077.9 |
PNKD
|
PNKD metallo-beta-lactamase domain containing |
| chr12_-_16608183 | 0.19 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr11_-_129192198 | 0.19 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr16_+_11965193 | 0.19 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr1_+_180632001 | 0.19 |
ENST00000367590.9
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr13_-_103066411 | 0.19 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2 |
| chr5_+_127649018 | 0.19 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr19_-_3557563 | 0.18 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr12_+_28257195 | 0.18 |
ENST00000381259.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_244352627 | 0.18 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr20_+_43916142 | 0.18 |
ENST00000423191.6
ENST00000372999.5 |
TOX2
|
TOX high mobility group box family member 2 |
| chr8_+_106726012 | 0.18 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr1_+_209704836 | 0.18 |
ENST00000367027.5
|
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr12_-_16608073 | 0.18 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr2_+_90038848 | 0.17 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr10_-_1737516 | 0.17 |
ENST00000381312.6
|
ADARB2
|
adenosine deaminase RNA specific B2 (inactive) |
| chr3_+_120908072 | 0.17 |
ENST00000273666.10
ENST00000471454.6 ENST00000472879.5 ENST00000497029.5 ENST00000492541.5 |
STXBP5L
|
syntaxin binding protein 5 like |
| chr4_+_70334963 | 0.17 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr7_-_111392915 | 0.17 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr10_+_116324440 | 0.17 |
ENST00000333254.4
|
CCDC172
|
coiled-coil domain containing 172 |
| chr2_-_49974155 | 0.17 |
ENST00000635519.1
|
NRXN1
|
neurexin 1 |
| chr1_-_92486916 | 0.17 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr3_-_98517096 | 0.16 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr14_+_22202561 | 0.16 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr1_+_65992389 | 0.16 |
ENST00000423207.6
|
PDE4B
|
phosphodiesterase 4B |
| chr7_+_130344837 | 0.16 |
ENST00000485477.5
ENST00000431780.6 |
CPA5
|
carboxypeptidase A5 |
| chr6_-_32190170 | 0.16 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2 |
| chr12_+_20810698 | 0.16 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr2_+_147845020 | 0.15 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A |
| chr8_+_7881387 | 0.15 |
ENST00000314357.4
|
DEFB103A
|
defensin beta 103A |
| chr2_+_186694007 | 0.15 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr7_+_116222804 | 0.14 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr12_+_26195647 | 0.14 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr12_-_86256267 | 0.14 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr3_-_108529322 | 0.14 |
ENST00000273353.4
|
MYH15
|
myosin heavy chain 15 |
| chr19_+_12938598 | 0.14 |
ENST00000586760.2
ENST00000316448.10 ENST00000588454.6 |
CALR
|
calreticulin |
| chrX_+_69616067 | 0.14 |
ENST00000338901.4
ENST00000525810.5 ENST00000527388.5 ENST00000374553.6 ENST00000374552.9 ENST00000524573.5 |
EDA
|
ectodysplasin A |
| chr21_-_30166782 | 0.14 |
ENST00000286808.5
|
CLDN17
|
claudin 17 |
| chr1_-_150765735 | 0.14 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr7_-_4862015 | 0.14 |
ENST00000404991.2
|
PAPOLB
|
poly(A) polymerase beta |
| chr15_+_58138368 | 0.14 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr14_+_22271921 | 0.13 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr19_+_44891206 | 0.13 |
ENST00000405636.6
ENST00000252487.9 ENST00000592434.5 ENST00000589649.1 ENST00000426677.7 |
TOMM40
|
translocase of outer mitochondrial membrane 40 |
| chr4_+_87650277 | 0.13 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr12_-_56934403 | 0.13 |
ENST00000293502.2
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C member 7 |
| chr2_+_102418642 | 0.13 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr11_-_117876612 | 0.13 |
ENST00000584230.1
ENST00000526014.6 ENST00000584394.5 ENST00000614497.5 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr8_-_7430348 | 0.13 |
ENST00000318124.3
|
DEFB103B
|
defensin beta 103B |
| chr11_-_57324907 | 0.13 |
ENST00000358252.8
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr12_+_80707625 | 0.13 |
ENST00000228641.4
|
MYF6
|
myogenic factor 6 |
| chr8_+_49911604 | 0.13 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr11_+_107591077 | 0.13 |
ENST00000531234.5
ENST00000265840.12 |
ELMOD1
|
ELMO domain containing 1 |
| chr1_+_84301694 | 0.12 |
ENST00000394834.8
ENST00000370669.5 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr14_+_22304051 | 0.12 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr8_+_49911396 | 0.12 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr6_-_37697875 | 0.12 |
ENST00000434837.8
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chrX_-_20218941 | 0.12 |
ENST00000457145.6
|
RPS6KA3
|
ribosomal protein S6 kinase A3 |
| chr18_+_78979811 | 0.12 |
ENST00000537592.7
|
SALL3
|
spalt like transcription factor 3 |
| chr8_+_106726115 | 0.12 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr13_-_94479671 | 0.12 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr4_+_87608529 | 0.12 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein |
| chr1_-_150765785 | 0.12 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr4_+_41612702 | 0.11 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr13_-_35855627 | 0.11 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SHOX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.4 | 1.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.3 | 1.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 0.6 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 | 0.7 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.2 | 0.7 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 0.9 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.6 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 1.7 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 1.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.5 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.3 | GO:0060503 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 2.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 3.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.2 | GO:0002668 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 0.2 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.4 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.6 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.2 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.4 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.5 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.5 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.8 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 2.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.3 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0042713 | sperm ejaculation(GO:0042713) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1902725 | regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 1.5 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 0.7 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.6 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 1.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 1.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 2.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.6 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 2.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 2.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 4.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |