Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for SOX13_SOX12
Z-value: 0.73
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
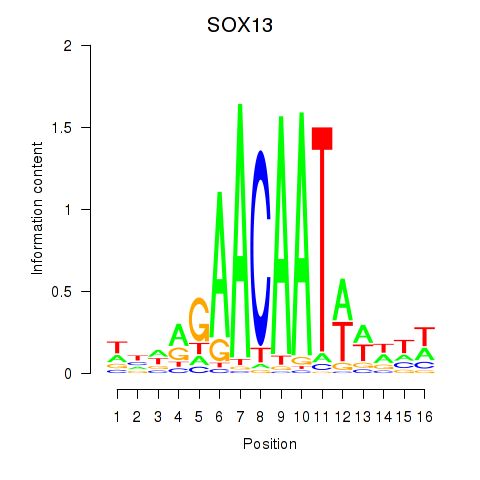
SOX13
|
ENSG00000143842.15 | SRY-box transcription factor 13 |
|
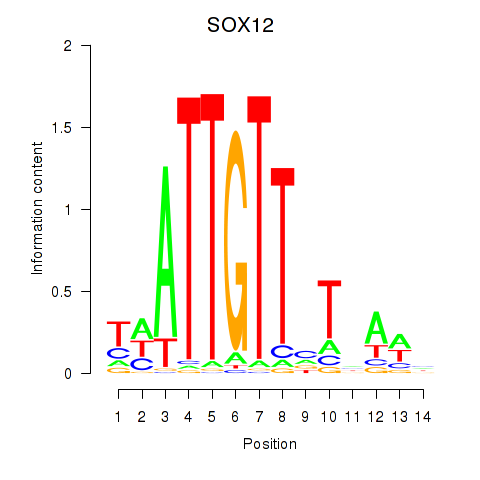
SOX12
|
ENSG00000177732.9 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX12 | hg38_v1_chr20_+_325536_325565 | -0.39 | 3.4e-01 | Click! |
| SOX13 | hg38_v1_chr1_+_204073104_204073121 | 0.20 | 6.3e-01 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_116427839 | 1.69 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr1_+_153031195 | 1.32 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B |
| chr12_-_8662703 | 0.94 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr12_-_8662619 | 0.88 |
ENST00000544889.1
ENST00000543369.5 |
MFAP5
|
microfibril associated protein 5 |
| chr6_-_136526472 | 0.82 |
ENST00000454590.5
ENST00000432797.6 |
MAP7
|
microtubule associated protein 7 |
| chr10_-_104085847 | 0.79 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr2_+_206939515 | 0.72 |
ENST00000272852.4
|
CPO
|
carboxypeptidase O |
| chr2_+_102311502 | 0.71 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr12_-_8650529 | 0.69 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr5_+_140841183 | 0.68 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr11_+_7088991 | 0.62 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr13_-_85799400 | 0.60 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr6_-_136466858 | 0.60 |
ENST00000544465.5
|
MAP7
|
microtubule associated protein 7 |
| chr12_-_8662808 | 0.60 |
ENST00000359478.7
ENST00000396549.6 |
MFAP5
|
microfibril associated protein 5 |
| chr19_+_35118456 | 0.58 |
ENST00000604621.5
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr3_-_151316795 | 0.55 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_+_140855882 | 0.55 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr12_+_50924005 | 0.53 |
ENST00000550502.1
|
METTL7A
|
methyltransferase like 7A |
| chr12_-_8662073 | 0.53 |
ENST00000535411.5
ENST00000540087.5 |
MFAP5
|
microfibril associated protein 5 |
| chr5_+_140834230 | 0.52 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr18_+_63587297 | 0.47 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr21_+_42199686 | 0.47 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr5_+_148203024 | 0.46 |
ENST00000325630.3
|
SPINK6
|
serine peptidase inhibitor Kazal type 6 |
| chr5_+_35856883 | 0.46 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr2_-_216013517 | 0.45 |
ENST00000263268.11
|
MREG
|
melanoregulin |
| chr18_+_63587336 | 0.45 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13 |
| chr1_+_76867469 | 0.44 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr1_-_153460644 | 0.43 |
ENST00000368723.4
|
S100A7
|
S100 calcium binding protein A7 |
| chr8_+_32721823 | 0.43 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr5_-_83673544 | 0.42 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_+_140868945 | 0.42 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11 |
| chr6_-_11778781 | 0.41 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr3_+_111998739 | 0.41 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr2_-_237590660 | 0.40 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr8_-_124728273 | 0.40 |
ENST00000325064.9
ENST00000518547.6 |
MTSS1
|
MTSS I-BAR domain containing 1 |
| chr5_+_36596583 | 0.39 |
ENST00000680318.1
|
SLC1A3
|
solute carrier family 1 member 3 |
| chr11_-_102780620 | 0.39 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr18_+_49562049 | 0.38 |
ENST00000261292.9
ENST00000427224.6 ENST00000580036.5 |
LIPG
|
lipase G, endothelial type |
| chr5_+_140882116 | 0.38 |
ENST00000289272.3
ENST00000409494.5 ENST00000617769.1 |
PCDHA13
|
protocadherin alpha 13 |
| chr12_+_8950036 | 0.38 |
ENST00000539240.5
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr12_-_27972725 | 0.37 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr5_+_148202771 | 0.36 |
ENST00000514389.5
ENST00000621437.4 |
SPINK6
|
serine peptidase inhibitor Kazal type 6 |
| chr6_-_136526654 | 0.36 |
ENST00000611373.1
|
MAP7
|
microtubule associated protein 7 |
| chr4_+_52862308 | 0.35 |
ENST00000248706.5
|
RASL11B
|
RAS like family 11 member B |
| chr18_+_63775369 | 0.33 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr2_+_131011683 | 0.33 |
ENST00000355771.7
|
ARHGEF4
|
Rho guanine nucleotide exchange factor 4 |
| chr11_-_60183011 | 0.32 |
ENST00000533023.5
ENST00000420732.6 ENST00000528851.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr18_+_58341038 | 0.32 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr5_+_140875299 | 0.32 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chr10_-_5003850 | 0.31 |
ENST00000421196.7
ENST00000455190.2 ENST00000380753.8 |
AKR1C2
|
aldo-keto reductase family 1 member C2 |
| chr16_+_2817230 | 0.31 |
ENST00000005995.8
ENST00000574813.5 |
PRSS21
|
serine protease 21 |
| chr11_+_128694052 | 0.31 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr9_-_137302264 | 0.31 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein |
| chr7_-_105522264 | 0.31 |
ENST00000469408.6
|
PUS7
|
pseudouridine synthase 7 |
| chr11_+_128693887 | 0.30 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_53510445 | 0.30 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chr2_-_237590694 | 0.30 |
ENST00000264601.8
ENST00000411462.5 ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr1_+_161707244 | 0.30 |
ENST00000349527.8
ENST00000294796.8 ENST00000309691.10 ENST00000367953.7 ENST00000367950.2 |
FCRLA
|
Fc receptor like A |
| chr2_-_70553440 | 0.29 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr6_+_125219804 | 0.29 |
ENST00000524679.1
|
TPD52L1
|
TPD52 like 1 |
| chr3_-_139539577 | 0.28 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1 |
| chr2_-_216013582 | 0.28 |
ENST00000620139.4
|
MREG
|
melanoregulin |
| chr6_+_130421086 | 0.28 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A |
| chr15_-_78937198 | 0.27 |
ENST00000677207.1
|
CTSH
|
cathepsin H |
| chr8_-_42501224 | 0.27 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr4_-_22443110 | 0.26 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr4_+_99816797 | 0.26 |
ENST00000512369.2
ENST00000296414.11 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr7_-_105522204 | 0.26 |
ENST00000356362.6
|
PUS7
|
pseudouridine synthase 7 |
| chr10_-_121598412 | 0.25 |
ENST00000360144.7
ENST00000358487.10 ENST00000369059.5 ENST00000613048.4 ENST00000356226.8 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr12_+_77830886 | 0.25 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chrX_+_108044967 | 0.25 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_+_102311546 | 0.24 |
ENST00000233954.6
ENST00000447231.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr11_+_55262152 | 0.24 |
ENST00000417545.5
|
TRIM48
|
tripartite motif containing 48 |
| chr3_-_112845950 | 0.24 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr7_-_41703062 | 0.24 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr4_+_76435216 | 0.23 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr10_-_114684612 | 0.23 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr15_-_99251207 | 0.23 |
ENST00000394129.6
ENST00000558663.5 ENST00000394135.7 ENST00000561365.5 ENST00000560279.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr7_+_117020191 | 0.23 |
ENST00000434836.5
ENST00000393443.5 ENST00000465133.5 ENST00000477742.5 ENST00000393444.7 ENST00000393447.8 |
ST7
|
suppression of tumorigenicity 7 |
| chr5_+_122129533 | 0.22 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr14_+_22271921 | 0.22 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr12_+_25052512 | 0.22 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr9_-_39239174 | 0.22 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr3_+_58008350 | 0.22 |
ENST00000490882.5
ENST00000358537.7 ENST00000429972.6 ENST00000682871.1 ENST00000295956.9 |
FLNB
|
filamin B |
| chr2_-_88947820 | 0.22 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr13_+_31739520 | 0.22 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr1_+_161706949 | 0.21 |
ENST00000350710.3
ENST00000367949.6 ENST00000367959.6 ENST00000540521.5 ENST00000546024.5 ENST00000674251.1 ENST00000674323.1 |
FCRLA
|
Fc receptor like A |
| chr1_-_153041111 | 0.21 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr12_-_84892120 | 0.21 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr6_+_106086316 | 0.21 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr4_-_121381007 | 0.21 |
ENST00000394427.3
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr1_+_161707205 | 0.21 |
ENST00000367957.7
|
FCRLA
|
Fc receptor like A |
| chr3_+_32391694 | 0.20 |
ENST00000349718.8
|
CMTM7
|
CKLF like MARVEL transmembrane domain containing 7 |
| chr1_+_161707222 | 0.20 |
ENST00000236938.12
|
FCRLA
|
Fc receptor like A |
| chr13_+_31739542 | 0.20 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr12_-_54384687 | 0.20 |
ENST00000550120.1
ENST00000547210.5 ENST00000394313.7 |
ZNF385A
|
zinc finger protein 385A |
| chr11_+_34632464 | 0.20 |
ENST00000531794.5
|
EHF
|
ETS homologous factor |
| chr13_+_73054969 | 0.20 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5 |
| chr9_-_21385395 | 0.20 |
ENST00000380206.4
|
IFNA2
|
interferon alpha 2 |
| chr4_+_99511008 | 0.20 |
ENST00000514652.5
ENST00000326581.9 |
C4orf17
|
chromosome 4 open reading frame 17 |
| chr19_-_55140922 | 0.20 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr10_-_14604389 | 0.19 |
ENST00000468747.5
ENST00000378467.8 |
FAM107B
|
family with sequence similarity 107 member B |
| chr20_-_51768327 | 0.19 |
ENST00000311637.9
ENST00000338821.6 |
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr11_+_35180342 | 0.19 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chrX_+_106693838 | 0.19 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr3_+_32391841 | 0.19 |
ENST00000334983.10
|
CMTM7
|
CKLF like MARVEL transmembrane domain containing 7 |
| chrX_+_108045050 | 0.19 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_130018565 | 0.19 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr1_-_153057504 | 0.18 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A |
| chr3_+_32391871 | 0.18 |
ENST00000465248.1
|
CMTM7
|
CKLF like MARVEL transmembrane domain containing 7 |
| chr17_+_58238426 | 0.18 |
ENST00000421678.6
ENST00000262290.9 ENST00000543544.5 |
LPO
|
lactoperoxidase |
| chr14_+_39233908 | 0.18 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_35176575 | 0.18 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr6_+_116399395 | 0.18 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr12_+_25052634 | 0.18 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr4_+_186227501 | 0.18 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr11_-_48983826 | 0.17 |
ENST00000649162.1
|
TRIM51GP
|
tripartite motif-containing 51G, pseudogene |
| chr6_+_137867414 | 0.17 |
ENST00000237289.8
ENST00000433680.1 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr2_+_90114838 | 0.17 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr19_-_42594918 | 0.17 |
ENST00000244336.10
|
CEACAM8
|
CEA cell adhesion molecule 8 |
| chr5_+_40841308 | 0.17 |
ENST00000381677.4
ENST00000254691.10 |
CARD6
|
caspase recruitment domain family member 6 |
| chr4_-_142305935 | 0.17 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr4_-_142305826 | 0.17 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr5_-_177409535 | 0.17 |
ENST00000253496.4
|
F12
|
coagulation factor XII |
| chr6_+_12290353 | 0.17 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chrX_-_32155462 | 0.17 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chr6_-_139291987 | 0.17 |
ENST00000358430.8
|
TXLNB
|
taxilin beta |
| chr11_-_89921767 | 0.17 |
ENST00000530311.6
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chrX_-_79367307 | 0.17 |
ENST00000373298.7
|
ITM2A
|
integral membrane protein 2A |
| chr8_-_85341705 | 0.17 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr6_-_53148822 | 0.17 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr6_-_49744434 | 0.16 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chrX_-_19965142 | 0.16 |
ENST00000340625.3
|
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chrX_+_23908006 | 0.16 |
ENST00000379211.8
ENST00000648352.1 |
CXorf58
|
chromosome X open reading frame 58 |
| chr17_+_55751021 | 0.16 |
ENST00000268896.10
ENST00000576183.5 ENST00000573500.5 |
PCTP
|
phosphatidylcholine transfer protein |
| chr18_+_44680875 | 0.16 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr5_+_141187138 | 0.15 |
ENST00000316105.7
ENST00000624909.1 |
PCDHB9
|
protocadherin beta 9 |
| chr10_-_59753388 | 0.15 |
ENST00000430431.5
|
MRLN
|
myoregulin |
| chr6_-_116060859 | 0.15 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr4_-_89838289 | 0.15 |
ENST00000336904.7
|
SNCA
|
synuclein alpha |
| chr14_+_56117702 | 0.15 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr10_-_107164692 | 0.15 |
ENST00000263054.11
|
SORCS1
|
sortilin related VPS10 domain containing receptor 1 |
| chr11_+_59713403 | 0.15 |
ENST00000641815.1
|
STX3
|
syntaxin 3 |
| chr1_-_152325232 | 0.15 |
ENST00000368799.2
|
FLG
|
filaggrin |
| chr19_+_4402615 | 0.15 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr7_-_36367141 | 0.15 |
ENST00000453212.5
ENST00000415803.2 ENST00000431396.1 ENST00000440378.6 ENST00000317020.10 ENST00000436884.5 |
KIAA0895
|
KIAA0895 |
| chr12_-_31326111 | 0.14 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr21_-_32813679 | 0.14 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr18_-_72544336 | 0.14 |
ENST00000269503.9
|
CBLN2
|
cerebellin 2 precursor |
| chr1_+_24319342 | 0.14 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr16_+_2817180 | 0.14 |
ENST00000450020.7
|
PRSS21
|
serine protease 21 |
| chr16_+_50266530 | 0.14 |
ENST00000566433.6
ENST00000394697.7 ENST00000673801.1 |
ADCY7
|
adenylate cyclase 7 |
| chr6_-_118935146 | 0.14 |
ENST00000619706.5
ENST00000316316.10 |
MCM9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr15_-_75712828 | 0.14 |
ENST00000308508.5
|
CSPG4
|
chondroitin sulfate proteoglycan 4 |
| chr11_-_60184633 | 0.14 |
ENST00000529054.5
ENST00000530839.5 ENST00000426738.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr9_-_33473884 | 0.14 |
ENST00000297990.9
ENST00000353159.6 ENST00000379471.3 |
NOL6
|
nucleolar protein 6 |
| chr19_+_40775154 | 0.14 |
ENST00000594436.5
ENST00000597784.5 |
MIA
|
MIA SH3 domain containing |
| chrX_+_105948429 | 0.14 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chrX_+_135309480 | 0.14 |
ENST00000635820.1
|
ETDC
|
embryonic testis differentiation homolog C |
| chr3_-_197226351 | 0.14 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr1_+_207752046 | 0.14 |
ENST00000367042.6
ENST00000322875.8 ENST00000322918.9 ENST00000354848.5 ENST00000357714.5 ENST00000358170.6 ENST00000367041.5 ENST00000367047.5 ENST00000360212.6 ENST00000480003.5 |
CD46
|
CD46 molecule |
| chr19_+_15628343 | 0.14 |
ENST00000589722.2
|
CYP4F8
|
cytochrome P450 family 4 subfamily F member 8 |
| chr19_+_1077394 | 0.14 |
ENST00000590577.2
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr15_-_79090760 | 0.13 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr6_+_89433164 | 0.13 |
ENST00000369408.9
ENST00000447838.6 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr10_-_23344678 | 0.13 |
ENST00000649478.1
ENST00000636213.3 ENST00000673651.1 ENST00000323327.5 |
C10orf67
|
chromosome 10 open reading frame 67 |
| chr19_+_21082140 | 0.13 |
ENST00000616183.1
ENST00000596053.5 |
ZNF714
|
zinc finger protein 714 |
| chr12_-_31326142 | 0.13 |
ENST00000337682.9
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr14_+_39233884 | 0.13 |
ENST00000553728.1
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_55811367 | 0.13 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr13_-_51974775 | 0.13 |
ENST00000674147.1
|
ATP7B
|
ATPase copper transporting beta |
| chr22_-_40862070 | 0.13 |
ENST00000307221.5
|
DNAJB7
|
DnaJ heat shock protein family (Hsp40) member B7 |
| chr1_+_153416517 | 0.13 |
ENST00000368729.9
|
S100A7A
|
S100 calcium binding protein A7A |
| chr6_-_49744378 | 0.13 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr12_-_31326171 | 0.13 |
ENST00000542983.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chrX_-_33211540 | 0.13 |
ENST00000357033.9
|
DMD
|
dystrophin |
| chr19_+_41708635 | 0.13 |
ENST00000617332.4
ENST00000615021.4 ENST00000616453.1 ENST00000405816.5 ENST00000435837.2 |
CEACAM5
ENSG00000267881.2
|
CEA cell adhesion molecule 5 novel protein, readthrough between CEACAM5-CEACAM6 |
| chr17_+_37491464 | 0.13 |
ENST00000613659.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr12_+_25052732 | 0.12 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr17_-_40665121 | 0.12 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr12_-_11134644 | 0.12 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr14_-_105864247 | 0.12 |
ENST00000461719.1
|
IGHJ4
|
immunoglobulin heavy joining 4 |
| chr9_-_123184233 | 0.12 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr13_-_46142834 | 0.12 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr8_+_102551583 | 0.12 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr21_-_26051023 | 0.12 |
ENST00000415997.1
|
APP
|
amyloid beta precursor protein |
| chr8_+_103819244 | 0.12 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_+_29395631 | 0.12 |
ENST00000642051.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr11_+_73950985 | 0.12 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr8_+_103880412 | 0.12 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr12_+_101594849 | 0.12 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr6_+_89433127 | 0.12 |
ENST00000339746.9
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr11_+_7605719 | 0.11 |
ENST00000530181.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr2_+_71453538 | 0.11 |
ENST00000258104.8
|
DYSF
|
dysferlin |
| chr12_-_23951020 | 0.11 |
ENST00000441133.2
ENST00000545921.5 |
SOX5
|
SRY-box transcription factor 5 |
| chr16_-_10694922 | 0.11 |
ENST00000283025.7
|
TEKT5
|
tektin 5 |
| chr12_+_48482492 | 0.11 |
ENST00000548364.7
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr4_+_122378966 | 0.11 |
ENST00000446706.5
ENST00000296513.7 |
ADAD1
|
adenosine deaminase domain containing 1 |
| chr7_+_77538027 | 0.11 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr6_-_112087451 | 0.11 |
ENST00000368662.10
|
TUBE1
|
tubulin epsilon 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 0.5 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 0.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 1.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.4 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.3 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.9 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.3 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 3.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.2 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.1 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 0.3 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:1905026 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.4 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.2 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.2 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0051563 | astrocyte activation involved in immune response(GO:0002265) microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 2.7 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of sodium:proton antiporter activity(GO:0032417) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 1.7 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.0 | GO:1900082 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0072387 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.0 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.0 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0032401 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 2.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.3 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 3.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0008903 | hydroxypyruvate isomerase activity(GO:0008903) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.0 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME DEVELOPMENTAL BIOLOGY | Genes involved in Developmental Biology |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |