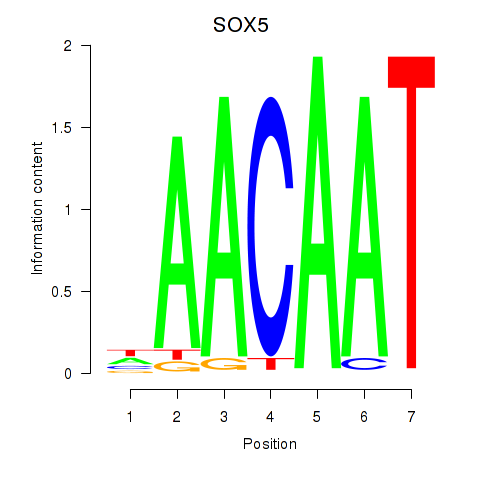
|
chr14_-_91947654
Show fit
|
0.93 |
ENST00000342058.9
|
FBLN5
|
fibulin 5
|
|
chr14_-_91947383
Show fit
|
0.89 |
ENST00000267620.14
|
FBLN5
|
fibulin 5
|
|
chr14_-_91946989
Show fit
|
0.83 |
ENST00000556154.5
|
FBLN5
|
fibulin 5
|
|
chr8_-_107497909
Show fit
|
0.65 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1
|
|
chr3_-_18424533
Show fit
|
0.55 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1
|
|
chr14_-_89417148
Show fit
|
0.48 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3
|
|
chr14_-_21536884
Show fit
|
0.44 |
ENST00000546363.5
|
SALL2
|
spalt like transcription factor 2
|
|
chr2_-_144431001
Show fit
|
0.44 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144430934
Show fit
|
0.43 |
ENST00000638087.1
ENST00000638007.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr5_-_159099684
Show fit
|
0.42 |
ENST00000380654.8
|
EBF1
|
EBF transcription factor 1
|
|
chr5_-_159099745
Show fit
|
0.41 |
ENST00000517373.1
|
EBF1
|
EBF transcription factor 1
|
|
chr4_-_2262082
Show fit
|
0.40 |
ENST00000337190.7
|
MXD4
|
MAX dimerization protein 4
|
|
chr5_-_159099909
Show fit
|
0.39 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1
|
|
chr5_-_168883333
Show fit
|
0.39 |
ENST00000404867.7
|
SLIT3
|
slit guidance ligand 3
|
|
chr10_+_68106109
Show fit
|
0.38 |
ENST00000540630.5
ENST00000354393.6
|
MYPN
|
myopalladin
|
|
chrX_-_107716401
Show fit
|
0.37 |
ENST00000486554.1
ENST00000372390.8
|
TSC22D3
|
TSC22 domain family member 3
|
|
chr12_+_53050014
Show fit
|
0.34 |
ENST00000314250.11
|
TNS2
|
tensin 2
|
|
chr12_+_53050179
Show fit
|
0.34 |
ENST00000546602.5
ENST00000552570.5
ENST00000549700.5
|
TNS2
|
tensin 2
|
|
chr3_-_115071333
Show fit
|
0.34 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_-_143635081
Show fit
|
0.32 |
ENST00000338017.8
|
SLITRK4
|
SLIT and NTRK like family member 4
|
|
chrX_+_136169891
Show fit
|
0.30 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_+_68292562
Show fit
|
0.28 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1
|
|
chrX_+_136169833
Show fit
|
0.28 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_+_136169664
Show fit
|
0.27 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_-_47582076
Show fit
|
0.26 |
ENST00000311160.14
|
TNS3
|
tensin 3
|
|
chr1_-_85578345
Show fit
|
0.26 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chrX_+_9912434
Show fit
|
0.26 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2
|
|
chr10_+_97319250
Show fit
|
0.25 |
ENST00000371021.5
|
FRAT1
|
FRAT regulator of WNT signaling pathway 1
|
|
chr1_+_164559739
Show fit
|
0.25 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1
|
|
chr2_-_199458689
Show fit
|
0.24 |
ENST00000443023.5
|
SATB2
|
SATB homeobox 2
|
|
chr6_+_89080739
Show fit
|
0.23 |
ENST00000369472.1
ENST00000336032.4
|
PNRC1
|
proline rich nuclear receptor coactivator 1
|
|
chr9_-_120477354
Show fit
|
0.23 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2
|
|
chr9_-_70869076
Show fit
|
0.23 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3
|
|
chr3_+_155079663
Show fit
|
0.22 |
ENST00000460393.6
|
MME
|
membrane metalloendopeptidase
|
|
chr14_-_73027077
Show fit
|
0.22 |
ENST00000553891.5
ENST00000556143.6
|
ZFYVE1
|
zinc finger FYVE-type containing 1
|
|
chr14_-_73027117
Show fit
|
0.22 |
ENST00000318876.9
|
ZFYVE1
|
zinc finger FYVE-type containing 1
|
|
chrX_+_136169624
Show fit
|
0.22 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1
|
|
chr15_+_43792839
Show fit
|
0.22 |
ENST00000409614.1
|
SERF2
|
small EDRK-rich factor 2
|
|
chr11_-_82997394
Show fit
|
0.21 |
ENST00000525117.5
ENST00000532548.5
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr10_-_32378720
Show fit
|
0.21 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1
|
|
chr20_-_47356670
Show fit
|
0.21 |
ENST00000540497.5
ENST00000461685.5
ENST00000617418.4
ENST00000435836.5
ENST00000471951.6
ENST00000352431.6
ENST00000360911.7
ENST00000458360.6
|
ZMYND8
|
zinc finger MYND-type containing 8
|
|
chr9_+_116153783
Show fit
|
0.21 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1
|
|
chr1_-_182391363
Show fit
|
0.20 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase
|
|
chrX_-_64976500
Show fit
|
0.20 |
ENST00000447788.6
|
ZC4H2
|
zinc finger C4H2-type containing
|
|
chr3_-_99876104
Show fit
|
0.20 |
ENST00000471562.1
ENST00000495625.2
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr20_-_47356721
Show fit
|
0.20 |
ENST00000262975.8
ENST00000446994.6
ENST00000355972.8
ENST00000396281.8
ENST00000619049.4
ENST00000611941.4
ENST00000372023.7
|
ZMYND8
|
zinc finger MYND-type containing 8
|
|
chr15_+_76336755
Show fit
|
0.20 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2
|
|
chr4_-_185810894
Show fit
|
0.20 |
ENST00000448662.6
ENST00000439049.5
ENST00000420158.5
ENST00000319471.13
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_156248013
Show fit
|
0.20 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6
|
|
chr1_+_164559766
Show fit
|
0.19 |
ENST00000367897.5
ENST00000559240.5
|
PBX1
|
PBX homeobox 1
|
|
chr7_-_27102669
Show fit
|
0.19 |
ENST00000222718.7
|
HOXA2
|
homeobox A2
|
|
chr5_+_139341826
Show fit
|
0.19 |
ENST00000265192.9
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr3_-_48717166
Show fit
|
0.18 |
ENST00000413654.5
ENST00000454335.5
ENST00000440424.5
ENST00000449610.5
ENST00000443964.1
ENST00000417896.1
ENST00000413298.5
ENST00000449563.5
ENST00000443853.5
ENST00000437427.5
ENST00000446860.5
ENST00000412850.5
ENST00000424035.1
ENST00000340879.8
ENST00000431721.6
ENST00000434860.1
ENST00000328631.10
ENST00000432678.6
|
IP6K2
|
inositol hexakisphosphate kinase 2
|
|
chr1_-_156248038
Show fit
|
0.18 |
ENST00000470198.5
ENST00000292291.10
ENST00000356983.7
|
PAQR6
|
progestin and adipoQ receptor family member 6
|
|
chr3_-_114624193
Show fit
|
0.18 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_-_156248084
Show fit
|
0.18 |
ENST00000652405.1
ENST00000335852.6
ENST00000540423.5
ENST00000612424.4
ENST00000613336.4
ENST00000623241.3
|
PAQR6
|
progestin and adipoQ receptor family member 6
|
|
chr22_+_37696982
Show fit
|
0.18 |
ENST00000644935.1
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr15_-_52652031
Show fit
|
0.18 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A
|
|
chr6_+_27810041
Show fit
|
0.17 |
ENST00000369163.3
|
H3C10
|
H3 clustered histone 10
|
|
chr11_-_82997477
Show fit
|
0.17 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr5_+_139341875
Show fit
|
0.17 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr17_+_69502397
Show fit
|
0.17 |
ENST00000613873.4
ENST00000589647.5
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr9_-_35684766
Show fit
|
0.17 |
ENST00000644325.1
|
TPM2
|
tropomyosin 2
|
|
chr2_-_2326378
Show fit
|
0.17 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr18_+_3451585
Show fit
|
0.17 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr20_-_18497218
Show fit
|
0.16 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase
|
|
chr13_-_74133892
Show fit
|
0.16 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12
|
|
chr6_+_155216637
Show fit
|
0.16 |
ENST00000275246.11
|
TIAM2
|
TIAM Rac1 associated GEF 2
|
|
chr9_-_15510954
Show fit
|
0.16 |
ENST00000380733.9
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr6_+_118894144
Show fit
|
0.16 |
ENST00000229595.6
|
ASF1A
|
anti-silencing function 1A histone chaperone
|
|
chr18_+_3451647
Show fit
|
0.15 |
ENST00000345133.9
ENST00000330513.10
ENST00000549546.5
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr14_-_50561119
Show fit
|
0.15 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5
|
|
chr17_-_16569184
Show fit
|
0.15 |
ENST00000448349.2
ENST00000395825.4
|
ZNF287
|
zinc finger protein 287
|
|
chrX_-_101617921
Show fit
|
0.15 |
ENST00000361910.9
ENST00000538627.5
ENST00000539247.5
|
ARMCX6
|
armadillo repeat containing X-linked 6
|
|
chr3_+_141386862
Show fit
|
0.15 |
ENST00000513258.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr3_-_71305986
Show fit
|
0.15 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1
|
|
chr5_+_173889337
Show fit
|
0.15 |
ENST00000520867.5
ENST00000334035.9
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr1_+_61081728
Show fit
|
0.14 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A
|
|
chr10_+_61901678
Show fit
|
0.14 |
ENST00000644638.1
ENST00000681100.1
ENST00000279873.12
|
ARID5B
|
AT-rich interaction domain 5B
|
|
chr9_-_70869029
Show fit
|
0.14 |
ENST00000361823.9
ENST00000377101.5
ENST00000360823.6
ENST00000377105.5
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3
|
|
chr12_-_31590967
Show fit
|
0.14 |
ENST00000354285.8
|
DENND5B
|
DENN domain containing 5B
|
|
chr17_-_64390852
Show fit
|
0.14 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1
|
|
chr2_-_110473041
Show fit
|
0.14 |
ENST00000632897.1
ENST00000413601.3
|
LIMS4
|
LIM zinc finger domain containing 4
|
|
chr5_-_88785493
Show fit
|
0.13 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_56851888
Show fit
|
0.13 |
ENST00000312431.10
ENST00000520645.5
|
DST
|
dystonin
|
|
chr16_+_86578543
Show fit
|
0.13 |
ENST00000320241.5
|
FOXL1
|
forkhead box L1
|
|
chr12_+_32502114
Show fit
|
0.13 |
ENST00000682739.1
ENST00000427716.7
ENST00000583694.2
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr20_+_17227020
Show fit
|
0.13 |
ENST00000262545.7
ENST00000536609.1
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr3_-_99876193
Show fit
|
0.13 |
ENST00000383694.3
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr10_+_102776237
Show fit
|
0.13 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like
|
|
chr1_-_36440873
Show fit
|
0.13 |
ENST00000433045.6
|
OSCP1
|
organic solute carrier partner 1
|
|
chr20_-_47355657
Show fit
|
0.13 |
ENST00000311275.11
|
ZMYND8
|
zinc finger MYND-type containing 8
|
|
chr8_-_71361860
Show fit
|
0.13 |
ENST00000303824.11
ENST00000645451.1
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr6_+_112236806
Show fit
|
0.13 |
ENST00000588837.5
ENST00000590293.5
ENST00000585450.5
ENST00000629766.2
ENST00000590804.5
ENST00000590584.4
ENST00000628122.2
ENST00000627025.1
ENST00000590673.5
ENST00000585611.5
ENST00000587816.2
|
LAMA4-AS1
ENSG00000281613.2
|
LAMA4 antisense RNA 1
novel ret finger protein-like 4B
|
|
chr7_+_134891566
Show fit
|
0.13 |
ENST00000424922.5
ENST00000495522.1
|
CALD1
|
caldesmon 1
|
|
chr1_+_33256479
Show fit
|
0.12 |
ENST00000539719.6
ENST00000483388.5
|
ZNF362
|
zinc finger protein 362
|
|
chr6_+_125919296
Show fit
|
0.12 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr10_+_24239181
Show fit
|
0.12 |
ENST00000438429.5
|
KIAA1217
|
KIAA1217
|
|
chr9_-_15510991
Show fit
|
0.12 |
ENST00000380715.5
ENST00000380716.8
ENST00000380738.8
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr10_+_35127162
Show fit
|
0.12 |
ENST00000354759.7
|
CREM
|
cAMP responsive element modulator
|
|
chr10_+_35126791
Show fit
|
0.12 |
ENST00000474362.5
ENST00000374721.7
|
CREM
|
cAMP responsive element modulator
|
|
chr1_+_226063466
Show fit
|
0.12 |
ENST00000666609.1
ENST00000661429.1
|
H3-3A
|
H3.3 histone A
|
|
chr6_+_125919210
Show fit
|
0.12 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr2_+_109898685
Show fit
|
0.12 |
ENST00000480744.2
|
LIMS3
|
LIM zinc finger domain containing 3
|
|
chr1_-_11691471
Show fit
|
0.11 |
ENST00000376672.5
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr8_-_71362054
Show fit
|
0.11 |
ENST00000340726.8
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr6_-_79234713
Show fit
|
0.11 |
ENST00000620514.1
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr3_+_141387616
Show fit
|
0.11 |
ENST00000509883.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr6_-_79234619
Show fit
|
0.11 |
ENST00000344726.9
ENST00000275036.11
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr4_-_75673360
Show fit
|
0.10 |
ENST00000676584.1
ENST00000678329.1
ENST00000677889.1
ENST00000357854.7
ENST00000359707.9
ENST00000676974.1
ENST00000678273.1
ENST00000677201.1
ENST00000678244.1
ENST00000677125.1
ENST00000677162.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr8_-_56211257
Show fit
|
0.10 |
ENST00000316981.8
ENST00000423799.6
ENST00000429357.2
|
PLAG1
|
PLAG1 zinc finger
|
|
chr1_-_11691608
Show fit
|
0.10 |
ENST00000376667.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr14_+_74348440
Show fit
|
0.10 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated
|
|
chr2_-_187448244
Show fit
|
0.10 |
ENST00000392370.8
ENST00000410068.5
ENST00000447403.5
ENST00000410102.5
|
CALCRL
|
calcitonin receptor like receptor
|
|
chr1_-_11691646
Show fit
|
0.10 |
ENST00000235310.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr5_-_38595396
Show fit
|
0.10 |
ENST00000263409.8
|
LIFR
|
LIF receptor subunit alpha
|
|
chr9_+_68780029
Show fit
|
0.10 |
ENST00000394264.7
|
PABIR1
|
PP2A Aalpha (PPP2R1A) and B55A (PPP2R2A) interacting phosphatase regulator 1
|
|
chr9_+_2159672
Show fit
|
0.10 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr17_+_35121609
Show fit
|
0.10 |
ENST00000158009.6
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr5_+_139342442
Show fit
|
0.10 |
ENST00000394795.6
ENST00000510080.1
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr14_+_23377136
Show fit
|
0.10 |
ENST00000382809.2
|
CMTM5
|
CKLF like MARVEL transmembrane domain containing 5
|
|
chr1_+_177170916
Show fit
|
0.10 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2
|
|
chr9_+_79573162
Show fit
|
0.10 |
ENST00000425506.5
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr10_+_122374685
Show fit
|
0.10 |
ENST00000368990.7
ENST00000368989.6
ENST00000463663.6
|
PLEKHA1
|
pleckstrin homology domain containing A1
|
|
chr3_-_57199938
Show fit
|
0.10 |
ENST00000473921.2
ENST00000295934.8
|
HESX1
|
HESX homeobox 1
|
|
chr7_+_134866831
Show fit
|
0.10 |
ENST00000435928.1
|
CALD1
|
caldesmon 1
|
|
chr11_+_111937320
Show fit
|
0.10 |
ENST00000440460.7
|
DIXDC1
|
DIX domain containing 1
|
|
chr9_-_127980976
Show fit
|
0.10 |
ENST00000373095.6
|
FAM102A
|
family with sequence similarity 102 member A
|
|
chr1_-_193186599
Show fit
|
0.09 |
ENST00000367434.5
|
B3GALT2
|
beta-1,3-galactosyltransferase 2
|
|
chrX_+_108439866
Show fit
|
0.09 |
ENST00000361603.7
|
COL4A5
|
collagen type IV alpha 5 chain
|
|
chrX_+_108439779
Show fit
|
0.09 |
ENST00000328300.11
|
COL4A5
|
collagen type IV alpha 5 chain
|
|
chr10_+_21524627
Show fit
|
0.09 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor
|
|
chr3_-_71306012
Show fit
|
0.09 |
ENST00000649431.1
ENST00000610810.5
|
FOXP1
|
forkhead box P1
|
|
chr22_-_35840218
Show fit
|
0.09 |
ENST00000414461.6
ENST00000416721.6
ENST00000449924.6
ENST00000262829.11
ENST00000397305.3
|
RBFOX2
|
RNA binding fox-1 homolog 2
|
|
chr15_-_37099306
Show fit
|
0.09 |
ENST00000557796.6
ENST00000397620.6
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_2331336
Show fit
|
0.09 |
ENST00000648933.1
ENST00000644820.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr9_-_127950716
Show fit
|
0.09 |
ENST00000373084.8
|
FAM102A
|
family with sequence similarity 102 member A
|
|
chr17_-_35121487
Show fit
|
0.09 |
ENST00000593039.5
|
ENSG00000267618.5
|
RAD51L3-RFFL readthrough
|
|
chr2_-_2331225
Show fit
|
0.09 |
ENST00000648627.1
ENST00000649663.1
ENST00000650560.1
ENST00000428368.7
ENST00000648316.1
ENST00000648665.1
ENST00000649313.1
ENST00000399161.7
ENST00000647738.2
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr4_+_159267737
Show fit
|
0.09 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2
|
|
chr6_+_36029082
Show fit
|
0.09 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14
|
|
chr7_+_28412511
Show fit
|
0.09 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_-_69833338
Show fit
|
0.09 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1
|
|
chr10_+_35127023
Show fit
|
0.09 |
ENST00000429130.7
ENST00000469949.6
ENST00000460270.5
|
CREM
|
cAMP responsive element modulator
|
|
chr11_+_126355634
Show fit
|
0.09 |
ENST00000227495.10
ENST00000676545.1
ENST00000678865.1
ENST00000444328.7
ENST00000677503.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4
|
|
chr14_-_55411817
Show fit
|
0.09 |
ENST00000247178.6
|
ATG14
|
autophagy related 14
|
|
chr16_+_1782940
Show fit
|
0.09 |
ENST00000262302.14
ENST00000563136.5
ENST00000565987.5
ENST00000568287.5
ENST00000565134.5
|
NUBP2
|
nucleotide binding protein 2
|
|
chr16_-_31074193
Show fit
|
0.08 |
ENST00000300849.5
|
ZNF668
|
zinc finger protein 668
|
|
chr2_+_119759875
Show fit
|
0.08 |
ENST00000263708.7
|
PTPN4
|
protein tyrosine phosphatase non-receptor type 4
|
|
chrX_-_108439472
Show fit
|
0.08 |
ENST00000372216.8
|
COL4A6
|
collagen type IV alpha 6 chain
|
|
chr7_+_134891400
Show fit
|
0.08 |
ENST00000393118.6
|
CALD1
|
caldesmon 1
|
|
chr16_+_2033264
Show fit
|
0.08 |
ENST00000565855.5
ENST00000566198.1
|
SLC9A3R2
|
SLC9A3 regulator 2
|
|
chr1_+_32651142
Show fit
|
0.08 |
ENST00000414241.7
|
RBBP4
|
RB binding protein 4, chromatin remodeling factor
|
|
chr12_+_59664677
Show fit
|
0.08 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7
|
|
chr16_+_3654683
Show fit
|
0.08 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1
|
|
chr3_-_142028617
Show fit
|
0.08 |
ENST00000477292.5
ENST00000478006.5
ENST00000495310.5
ENST00000486111.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr4_-_7939789
Show fit
|
0.08 |
ENST00000420658.6
ENST00000358461.6
|
AFAP1
|
actin filament associated protein 1
|
|
chr20_+_6007245
Show fit
|
0.08 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1
|
|
chr7_+_129375643
Show fit
|
0.08 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2
|
|
chr3_-_46863435
Show fit
|
0.08 |
ENST00000395869.5
ENST00000653454.1
ENST00000292327.6
|
MYL3
|
myosin light chain 3
|
|
chr3_+_89107649
Show fit
|
0.08 |
ENST00000452448.6
ENST00000494014.1
|
EPHA3
|
EPH receptor A3
|
|
chrX_-_13817027
Show fit
|
0.08 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr19_+_7516081
Show fit
|
0.08 |
ENST00000597229.2
|
ZNF358
|
zinc finger protein 358
|
|
chr7_-_7535863
Show fit
|
0.08 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain
|
|
chr19_+_12838437
Show fit
|
0.07 |
ENST00000251472.9
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr4_+_113049479
Show fit
|
0.07 |
ENST00000671727.1
ENST00000671762.1
ENST00000672366.1
ENST00000672502.1
ENST00000672045.1
ENST00000672251.1
ENST00000672854.1
|
ANK2
|
ankyrin 2
|
|
chr16_-_1782526
Show fit
|
0.07 |
ENST00000566339.6
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr4_-_75673139
Show fit
|
0.07 |
ENST00000677566.1
ENST00000503660.5
ENST00000677060.1
ENST00000678552.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr1_-_50423431
Show fit
|
0.07 |
ENST00000404795.4
|
DMRTA2
|
DMRT like family A2
|
|
chr3_-_142028597
Show fit
|
0.07 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr4_+_109827963
Show fit
|
0.07 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chr6_+_57046681
Show fit
|
0.07 |
ENST00000370733.5
|
KIAA1586
|
KIAA1586
|
|
chr14_+_36657560
Show fit
|
0.07 |
ENST00000402703.6
|
PAX9
|
paired box 9
|
|
chr10_+_21524670
Show fit
|
0.07 |
ENST00000631589.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor
|
|
chr9_-_95509241
Show fit
|
0.07 |
ENST00000331920.11
|
PTCH1
|
patched 1
|
|
chr4_-_75672868
Show fit
|
0.06 |
ENST00000678123.1
ENST00000678578.1
ENST00000677876.1
ENST00000676839.1
ENST00000678265.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr14_-_69153106
Show fit
|
0.06 |
ENST00000341516.10
|
DCAF5
|
DDB1 and CUL4 associated factor 5
|
|
chr19_-_1568301
Show fit
|
0.06 |
ENST00000402693.5
|
MEX3D
|
mex-3 RNA binding family member D
|
|
chr20_+_31968141
Show fit
|
0.06 |
ENST00000562532.3
|
XKR7
|
XK related 7
|
|
chr10_+_48306698
Show fit
|
0.06 |
ENST00000374179.8
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr9_-_120714457
Show fit
|
0.06 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9
|
|
chr10_+_35127295
Show fit
|
0.06 |
ENST00000489321.5
ENST00000427847.6
ENST00000374728.7
ENST00000345491.7
ENST00000487132.5
|
CREM
|
cAMP responsive element modulator
|
|
chr6_+_57046532
Show fit
|
0.06 |
ENST00000545356.5
|
KIAA1586
|
KIAA1586
|
|
chr2_-_208124514
Show fit
|
0.06 |
ENST00000264376.5
|
CRYGD
|
crystallin gamma D
|
|
chr14_+_22508602
Show fit
|
0.06 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33
|
|
chr4_-_75673112
Show fit
|
0.06 |
ENST00000395719.7
ENST00000677489.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr1_+_153259684
Show fit
|
0.06 |
ENST00000368742.4
|
LORICRIN
|
loricrin cornified envelope precursor protein
|
|
chr3_+_89107613
Show fit
|
0.06 |
ENST00000336596.7
|
EPHA3
|
EPH receptor A3
|
|
chr10_+_48306639
Show fit
|
0.06 |
ENST00000395611.7
ENST00000432379.5
ENST00000374189.5
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr6_-_43059367
Show fit
|
0.06 |
ENST00000230413.9
ENST00000487429.1
ENST00000388752.8
ENST00000489623.1
ENST00000468957.1
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr8_+_56211686
Show fit
|
0.06 |
ENST00000521831.5
ENST00000303759.3
ENST00000517636.5
ENST00000517933.5
ENST00000355315.8
ENST00000518801.5
ENST00000523975.5
ENST00000396723.9
ENST00000523061.5
ENST00000521524.5
|
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7
|
|
chr5_-_135535955
Show fit
|
0.06 |
ENST00000314744.6
|
NEUROG1
|
neurogenin 1
|
|
chr3_+_19947316
Show fit
|
0.06 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr3_-_71493500
Show fit
|
0.06 |
ENST00000648380.1
ENST00000650295.1
|
FOXP1
|
forkhead box P1
|
|
chr9_-_131270493
Show fit
|
0.06 |
ENST00000372269.7
ENST00000464831.1
|
FAM78A
|
family with sequence similarity 78 member A
|
|
chr7_+_117480011
Show fit
|
0.05 |
ENST00000649406.1
ENST00000648260.1
ENST00000003084.11
|
CFTR
|
CF transmembrane conductance regulator
|
|
chr1_+_8945858
Show fit
|
0.05 |
ENST00000549778.5
ENST00000377443.7
ENST00000480186.7
ENST00000377436.6
ENST00000377442.3
|
CA6
|
carbonic anhydrase 6
|
|
chr8_+_67952028
Show fit
|
0.05 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2
|
|
chr12_+_49367364
Show fit
|
0.05 |
ENST00000549298.5
|
SPATS2
|
spermatogenesis associated serine rich 2
|
|
chr12_+_49367440
Show fit
|
0.05 |
ENST00000552918.6
ENST00000553127.5
ENST00000321898.10
|
SPATS2
|
spermatogenesis associated serine rich 2
|