Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
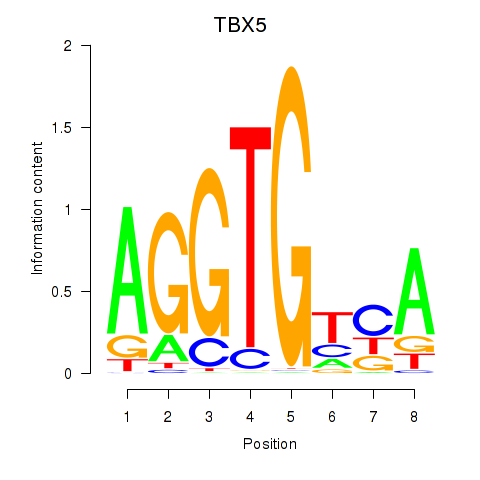
Results for TBX5
Z-value: 0.57
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.20 | T-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX5 | hg38_v1_chr12_-_114403898_114403921 | 0.55 | 1.6e-01 | Click! |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52493250 | 0.75 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr1_-_12616762 | 0.64 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr14_-_105168753 | 0.60 |
ENST00000331782.8
ENST00000347004.2 |
JAG2
|
jagged canonical Notch ligand 2 |
| chr2_+_68734773 | 0.60 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_+_101237009 | 0.57 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr16_-_31135699 | 0.50 |
ENST00000317508.11
ENST00000568261.5 ENST00000567797.1 |
PRSS8
|
serine protease 8 |
| chr8_+_32548303 | 0.47 |
ENST00000650967.1
|
NRG1
|
neuregulin 1 |
| chr1_+_2050387 | 0.44 |
ENST00000378567.8
|
PRKCZ
|
protein kinase C zeta |
| chr1_-_205321737 | 0.43 |
ENST00000367157.6
|
NUAK2
|
NUAK family kinase 2 |
| chr12_+_107318395 | 0.43 |
ENST00000420571.6
ENST00000280758.10 |
BTBD11
|
BTB domain containing 11 |
| chr8_+_32548210 | 0.43 |
ENST00000523079.5
ENST00000650919.1 |
NRG1
|
neuregulin 1 |
| chr3_-_134029914 | 0.41 |
ENST00000493729.5
ENST00000310926.11 |
SLCO2A1
|
solute carrier organic anion transporter family member 2A1 |
| chr2_-_215138603 | 0.40 |
ENST00000272895.12
|
ABCA12
|
ATP binding cassette subfamily A member 12 |
| chr15_+_40239857 | 0.38 |
ENST00000260404.8
|
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr14_+_75280078 | 0.37 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr1_+_59814939 | 0.37 |
ENST00000371208.5
|
HOOK1
|
hook microtubule tethering protein 1 |
| chr12_-_27971970 | 0.36 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr5_+_66828762 | 0.34 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_+_21057822 | 0.33 |
ENST00000308227.2
|
RNASE8
|
ribonuclease A family member 8 |
| chr8_-_74321532 | 0.33 |
ENST00000342232.5
|
JPH1
|
junctophilin 1 |
| chr6_-_13487593 | 0.32 |
ENST00000379287.4
ENST00000603223.1 |
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr18_-_55302613 | 0.31 |
ENST00000561831.7
|
TCF4
|
transcription factor 4 |
| chr8_+_32548267 | 0.30 |
ENST00000356819.7
|
NRG1
|
neuregulin 1 |
| chr1_+_151511376 | 0.30 |
ENST00000427934.2
ENST00000271636.12 |
CGN
|
cingulin |
| chr2_+_68734861 | 0.30 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr8_-_22131043 | 0.27 |
ENST00000312841.9
|
HR
|
HR lysine demethylase and nuclear receptor corepressor |
| chr8_+_119208322 | 0.27 |
ENST00000614891.5
|
MAL2
|
mal, T cell differentiation protein 2 |
| chr11_+_1838970 | 0.26 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type |
| chr8_-_22131003 | 0.26 |
ENST00000381418.9
|
HR
|
HR lysine demethylase and nuclear receptor corepressor |
| chr3_+_167735704 | 0.26 |
ENST00000446050.7
ENST00000295777.9 ENST00000472747.2 |
SERPINI1
|
serpin family I member 1 |
| chr12_-_52434363 | 0.26 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr14_+_61529005 | 0.25 |
ENST00000556347.1
|
ENSG00000258989.1
|
novel protein |
| chr2_-_162318129 | 0.25 |
ENST00000679938.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr1_+_13584262 | 0.25 |
ENST00000376061.8
ENST00000513143.5 |
PDPN
|
podoplanin |
| chr11_-_16356538 | 0.24 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr1_+_13583762 | 0.24 |
ENST00000376057.8
ENST00000621990.5 ENST00000510906.5 |
PDPN
|
podoplanin |
| chr13_+_77535669 | 0.24 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr14_+_21042352 | 0.23 |
ENST00000298690.5
|
RNASE7
|
ribonuclease A family member 7 |
| chr17_+_49495286 | 0.23 |
ENST00000172229.8
|
NGFR
|
nerve growth factor receptor |
| chr1_+_156284299 | 0.23 |
ENST00000456810.1
ENST00000405535.3 |
TMEM79
|
transmembrane protein 79 |
| chr13_+_77535681 | 0.23 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535742 | 0.22 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr8_+_122781621 | 0.22 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr14_+_75278820 | 0.22 |
ENST00000554617.1
ENST00000554212.5 ENST00000535987.5 ENST00000303562.9 ENST00000555242.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr8_+_31639291 | 0.21 |
ENST00000651149.1
ENST00000650866.1 |
NRG1
|
neuregulin 1 |
| chr15_+_88639009 | 0.21 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr8_+_31639222 | 0.21 |
ENST00000519301.6
ENST00000652698.1 |
NRG1
|
neuregulin 1 |
| chr6_-_2841853 | 0.20 |
ENST00000380739.6
|
SERPINB1
|
serpin family B member 1 |
| chr9_-_133149334 | 0.19 |
ENST00000393160.7
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr11_+_35176575 | 0.19 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr15_+_89776326 | 0.18 |
ENST00000341735.5
|
MESP2
|
mesoderm posterior bHLH transcription factor 2 |
| chr17_+_32487686 | 0.18 |
ENST00000584792.5
|
CDK5R1
|
cyclin dependent kinase 5 regulatory subunit 1 |
| chr1_-_153549238 | 0.18 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chrX_+_136536099 | 0.18 |
ENST00000440515.5
ENST00000456412.1 |
VGLL1
|
vestigial like family member 1 |
| chr3_+_142596385 | 0.18 |
ENST00000457734.7
ENST00000483373.5 ENST00000475296.5 ENST00000495744.5 ENST00000476044.5 ENST00000461644.5 ENST00000464320.5 |
PLS1
|
plastin 1 |
| chr4_+_119212636 | 0.18 |
ENST00000274030.10
|
USP53
|
ubiquitin specific peptidase 53 |
| chr3_-_183162726 | 0.17 |
ENST00000265598.8
|
LAMP3
|
lysosomal associated membrane protein 3 |
| chr8_+_127737610 | 0.17 |
ENST00000652288.1
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr15_+_88638947 | 0.17 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr12_+_19205294 | 0.17 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr21_+_42513834 | 0.17 |
ENST00000352133.3
|
SLC37A1
|
solute carrier family 37 member 1 |
| chrX_-_129654946 | 0.17 |
ENST00000429967.3
|
APLN
|
apelin |
| chr1_-_156816841 | 0.16 |
ENST00000368199.8
ENST00000392306.2 |
SH2D2A
|
SH2 domain containing 2A |
| chr18_+_32018817 | 0.16 |
ENST00000217740.4
ENST00000583184.1 |
RNF125
ENSG00000263917.1
|
ring finger protein 125 novel transcript |
| chr11_-_58578096 | 0.16 |
ENST00000528954.5
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr8_-_143572748 | 0.16 |
ENST00000529971.1
ENST00000398882.8 |
MROH6
|
maestro heat like repeat family member 6 |
| chr5_-_177409535 | 0.16 |
ENST00000253496.4
|
F12
|
coagulation factor XII |
| chr10_-_124093582 | 0.16 |
ENST00000462406.1
ENST00000435907.6 |
CHST15
|
carbohydrate sulfotransferase 15 |
| chr11_+_67056805 | 0.16 |
ENST00000308831.7
|
RHOD
|
ras homolog family member D |
| chr3_-_129121761 | 0.16 |
ENST00000476465.5
ENST00000393304.5 ENST00000315150.10 ENST00000615093.1 ENST00000393308.5 ENST00000393307.5 ENST00000393305.5 |
RAB43
|
RAB43, member RAS oncogene family |
| chr1_-_206921867 | 0.16 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr2_+_152335163 | 0.15 |
ENST00000288670.14
|
FMNL2
|
formin like 2 |
| chr1_-_156816738 | 0.15 |
ENST00000368198.7
|
SH2D2A
|
SH2 domain containing 2A |
| chr21_+_42219123 | 0.15 |
ENST00000398449.8
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr8_+_103819244 | 0.15 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr17_+_57105899 | 0.15 |
ENST00000576295.5
|
AKAP1
|
A-kinase anchoring protein 1 |
| chr1_+_153774210 | 0.15 |
ENST00000271857.6
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr20_-_22584547 | 0.15 |
ENST00000419308.7
|
FOXA2
|
forkhead box A2 |
| chr2_+_170816868 | 0.14 |
ENST00000358196.8
|
GAD1
|
glutamate decarboxylase 1 |
| chr1_-_54406385 | 0.14 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr1_-_204359885 | 0.14 |
ENST00000414478.1
ENST00000272203.8 |
PLEKHA6
|
pleckstrin homology domain containing A6 |
| chr17_+_39737923 | 0.14 |
ENST00000577695.5
ENST00000309156.9 |
GRB7
|
growth factor receptor bound protein 7 |
| chr17_-_28902930 | 0.14 |
ENST00000426464.2
|
DHRS13
|
dehydrogenase/reductase 13 |
| chr21_+_42219111 | 0.14 |
ENST00000450121.5
ENST00000361802.6 |
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr12_-_120904337 | 0.14 |
ENST00000353487.7
|
SPPL3
|
signal peptide peptidase like 3 |
| chr16_-_69351778 | 0.14 |
ENST00000288025.4
|
TMED6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_74529670 | 0.14 |
ENST00000377526.4
|
AUP1
|
AUP1 lipid droplet regulating VLDL assembly factor |
| chr5_-_135578983 | 0.13 |
ENST00000512158.6
|
CXCL14
|
C-X-C motif chemokine ligand 14 |
| chrX_+_21374476 | 0.13 |
ENST00000644585.1
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr2_-_74441882 | 0.13 |
ENST00000272430.10
|
RTKN
|
rhotekin |
| chr17_+_36103819 | 0.13 |
ENST00000615863.2
ENST00000621626.1 |
CCL4
|
C-C motif chemokine ligand 4 |
| chr11_-_34513750 | 0.13 |
ENST00000532417.1
|
ELF5
|
E74 like ETS transcription factor 5 |
| chrX_+_16786421 | 0.13 |
ENST00000398155.4
ENST00000380122.10 |
TXLNG
|
taxilin gamma |
| chr7_-_27095972 | 0.13 |
ENST00000355633.5
ENST00000643460.2 |
HOXA1
|
homeobox A1 |
| chr13_+_24270681 | 0.13 |
ENST00000343003.10
ENST00000399949.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr11_+_67056875 | 0.12 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr9_-_93955347 | 0.12 |
ENST00000253968.11
|
BARX1
|
BARX homeobox 1 |
| chr3_+_50267606 | 0.12 |
ENST00000618865.4
|
SEMA3B
|
semaphorin 3B |
| chr8_-_24956604 | 0.12 |
ENST00000610854.2
|
NEFL
|
neurofilament light |
| chr1_+_206507520 | 0.12 |
ENST00000579436.7
|
RASSF5
|
Ras association domain family member 5 |
| chr16_+_2988256 | 0.12 |
ENST00000573315.2
|
LINC00514
|
long intergenic non-protein coding RNA 514 |
| chr3_+_49689531 | 0.12 |
ENST00000432042.5
ENST00000454491.5 ENST00000327697.11 |
RNF123
|
ring finger protein 123 |
| chr2_-_72147819 | 0.12 |
ENST00000001146.7
ENST00000546307.5 ENST00000474509.1 |
CYP26B1
|
cytochrome P450 family 26 subfamily B member 1 |
| chr17_-_76537699 | 0.12 |
ENST00000293230.10
|
CYGB
|
cytoglobin |
| chr17_-_28903017 | 0.12 |
ENST00000394901.7
ENST00000378895.9 |
DHRS13
|
dehydrogenase/reductase 13 |
| chr1_-_202808406 | 0.11 |
ENST00000650569.1
ENST00000367265.9 ENST00000649770.1 |
KDM5B
|
lysine demethylase 5B |
| chr7_-_138981307 | 0.11 |
ENST00000440172.5
ENST00000422774.2 |
KIAA1549
|
KIAA1549 |
| chr11_+_19117123 | 0.11 |
ENST00000399351.7
ENST00000446113.7 |
ZDHHC13
|
zinc finger DHHC-type palmitoyltransferase 13 |
| chr7_-_100656384 | 0.11 |
ENST00000461605.1
ENST00000160382.10 |
ACTL6B
|
actin like 6B |
| chr3_-_52409783 | 0.11 |
ENST00000470173.1
ENST00000296288.9 ENST00000460680.6 |
BAP1
|
BRCA1 associated protein 1 |
| chr5_+_139795795 | 0.11 |
ENST00000274710.4
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr5_-_54985579 | 0.11 |
ENST00000381405.5
ENST00000381403.4 |
ESM1
|
endothelial cell specific molecule 1 |
| chr6_+_30326835 | 0.10 |
ENST00000440271.5
ENST00000376656.8 ENST00000396551.7 ENST00000428728.5 ENST00000396548.5 ENST00000428404.5 |
TRIM39
|
tripartite motif containing 39 |
| chr18_-_31162849 | 0.10 |
ENST00000257197.7
ENST00000257198.6 |
DSC1
|
desmocollin 1 |
| chr17_-_4560564 | 0.10 |
ENST00000574584.1
ENST00000381550.8 ENST00000301395.7 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr11_+_35176611 | 0.10 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_34513785 | 0.10 |
ENST00000257832.7
ENST00000429939.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr11_-_72674394 | 0.10 |
ENST00000418754.6
ENST00000334456.10 ENST00000542969.2 |
PDE2A
|
phosphodiesterase 2A |
| chr4_-_40629842 | 0.10 |
ENST00000295971.12
|
RBM47
|
RNA binding motif protein 47 |
| chr2_+_11556337 | 0.09 |
ENST00000234142.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr4_-_109729956 | 0.09 |
ENST00000502283.1
|
PLA2G12A
|
phospholipase A2 group XIIA |
| chr11_-_59615673 | 0.09 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein |
| chr11_-_123654581 | 0.09 |
ENST00000392770.6
ENST00000530277.5 ENST00000299333.8 |
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr1_-_10982037 | 0.09 |
ENST00000377004.8
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr3_+_4680617 | 0.09 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr11_-_67647577 | 0.09 |
ENST00000529256.1
|
ACY3
|
aminoacylase 3 |
| chr10_-_124092445 | 0.09 |
ENST00000346248.7
|
CHST15
|
carbohydrate sulfotransferase 15 |
| chr17_+_37489882 | 0.09 |
ENST00000617516.5
|
DUSP14
|
dual specificity phosphatase 14 |
| chr17_+_76376581 | 0.09 |
ENST00000591651.5
ENST00000545180.5 |
SPHK1
|
sphingosine kinase 1 |
| chr7_-_80919017 | 0.09 |
ENST00000265361.8
|
SEMA3C
|
semaphorin 3C |
| chr11_-_128867364 | 0.09 |
ENST00000440599.6
ENST00000324036.7 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chrX_+_21374357 | 0.09 |
ENST00000643841.1
ENST00000379510.5 ENST00000425654.7 ENST00000644798.1 ENST00000543067.6 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr13_-_103066411 | 0.09 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2 |
| chr6_+_25726767 | 0.09 |
ENST00000274764.5
|
H2BC1
|
H2B clustered histone 1 |
| chr11_-_128867268 | 0.09 |
ENST00000392665.6
ENST00000392666.6 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr1_+_165543992 | 0.09 |
ENST00000294818.2
|
LRRC52
|
leucine rich repeat containing 52 |
| chr11_-_132943671 | 0.09 |
ENST00000331898.11
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr20_+_34363241 | 0.09 |
ENST00000486883.5
ENST00000374864.10 ENST00000535650.7 ENST00000670516.1 ENST00000665484.1 ENST00000654846.1 ENST00000665428.1 ENST00000660337.1 ENST00000262650.10 ENST00000665346.1 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr21_-_45542465 | 0.08 |
ENST00000380010.8
|
SLC19A1
|
solute carrier family 19 member 1 |
| chr1_+_26863140 | 0.08 |
ENST00000339276.6
|
SFN
|
stratifin |
| chr12_-_18738006 | 0.08 |
ENST00000266505.12
ENST00000543242.5 ENST00000539072.5 ENST00000541966.5 ENST00000648272.1 |
PLCZ1
|
phospholipase C zeta 1 |
| chr1_+_15756659 | 0.08 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1 |
| chrX_-_110318062 | 0.08 |
ENST00000372059.6
ENST00000262844.10 |
AMMECR1
|
AMMECR nuclear protein 1 |
| chr12_+_80707625 | 0.08 |
ENST00000228641.4
|
MYF6
|
myogenic factor 6 |
| chr16_-_11256192 | 0.08 |
ENST00000644787.1
ENST00000332029.4 |
SOCS1
|
suppressor of cytokine signaling 1 |
| chr1_+_7784411 | 0.08 |
ENST00000613533.4
ENST00000614998.4 |
PER3
|
period circadian regulator 3 |
| chr8_-_20303955 | 0.08 |
ENST00000381569.5
|
LZTS1
|
leucine zipper tumor suppressor 1 |
| chr1_+_93079264 | 0.08 |
ENST00000370298.9
ENST00000370303.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr1_-_44017296 | 0.08 |
ENST00000357730.6
ENST00000360584.6 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr4_-_151015713 | 0.08 |
ENST00000357115.9
|
LRBA
|
LPS responsive beige-like anchor protein |
| chr2_+_29113989 | 0.08 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr18_-_3874247 | 0.08 |
ENST00000581699.5
|
DLGAP1
|
DLG associated protein 1 |
| chr3_+_184315347 | 0.07 |
ENST00000424196.5
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr19_-_41428730 | 0.07 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr1_+_93079234 | 0.07 |
ENST00000540243.5
ENST00000545708.5 |
MTF2
|
metal response element binding transcription factor 2 |
| chr1_+_158355894 | 0.07 |
ENST00000368162.2
|
CD1E
|
CD1e molecule |
| chr7_+_142511614 | 0.07 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr2_+_170715317 | 0.07 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor |
| chr11_-_95231046 | 0.07 |
ENST00000416495.6
ENST00000536441.7 |
SESN3
|
sestrin 3 |
| chr11_+_34622053 | 0.07 |
ENST00000530286.5
ENST00000533754.5 |
EHF
|
ETS homologous factor |
| chr1_-_11805924 | 0.07 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr4_+_80197493 | 0.07 |
ENST00000415738.3
|
PRDM8
|
PR/SET domain 8 |
| chrX_-_49200174 | 0.07 |
ENST00000472598.5
ENST00000263233.9 ENST00000479808.5 |
SYP
|
synaptophysin |
| chr19_+_10286971 | 0.07 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr19_+_10286944 | 0.07 |
ENST00000380770.5
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr19_+_41443130 | 0.07 |
ENST00000378187.3
|
ERICH4
|
glutamate rich 4 |
| chr6_+_30327259 | 0.07 |
ENST00000376659.9
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr12_+_30926716 | 0.07 |
ENST00000546076.6
ENST00000535215.5 ENST00000261177.10 |
TSPAN11
|
tetraspanin 11 |
| chr10_+_133527355 | 0.07 |
ENST00000252945.8
ENST00000421586.5 ENST00000418356.1 |
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1 |
| chr19_+_17751467 | 0.07 |
ENST00000596536.5
ENST00000593870.5 ENST00000598086.5 ENST00000598932.5 ENST00000595023.5 ENST00000594068.5 ENST00000596507.5 ENST00000595033.5 ENST00000597718.5 |
FCHO1
|
FCH and mu domain containing endocytic adaptor 1 |
| chr2_+_233760265 | 0.07 |
ENST00000305208.10
ENST00000360418.4 |
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1 |
| chr20_-_40689228 | 0.07 |
ENST00000373313.3
|
MAFB
|
MAF bZIP transcription factor B |
| chrX_-_55030970 | 0.07 |
ENST00000493869.2
ENST00000396198.7 ENST00000650242.1 ENST00000335854.8 ENST00000477869.6 ENST00000455688.2 ENST00000644983.1 |
ALAS2
|
5'-aminolevulinate synthase 2 |
| chr11_+_46332905 | 0.07 |
ENST00000343674.10
|
DGKZ
|
diacylglycerol kinase zeta |
| chr12_+_6946468 | 0.07 |
ENST00000543115.5
ENST00000399448.5 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr1_+_99646025 | 0.07 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr3_+_184315131 | 0.06 |
ENST00000427845.5
ENST00000342981.8 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chrX_+_21374434 | 0.06 |
ENST00000279451.9
ENST00000645245.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr22_+_31082860 | 0.06 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chrX_-_131289412 | 0.06 |
ENST00000652189.1
ENST00000651556.1 |
IGSF1
|
immunoglobulin superfamily member 1 |
| chr1_+_171781635 | 0.06 |
ENST00000361735.4
ENST00000362019.7 ENST00000367737.9 |
EEF1AKNMT
|
eEF1A lysine and N-terminal methyltransferase |
| chr7_-_82443766 | 0.06 |
ENST00000356860.8
|
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr17_+_17042433 | 0.06 |
ENST00000651222.2
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr16_+_2019777 | 0.06 |
ENST00000566435.4
|
NPW
|
neuropeptide W |
| chr7_-_82443715 | 0.06 |
ENST00000356253.9
ENST00000423588.1 |
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr10_-_101229449 | 0.06 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr13_-_35855627 | 0.06 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr10_+_113125536 | 0.06 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr16_+_727117 | 0.06 |
ENST00000562141.5
|
HAGHL
|
hydroxyacylglutathione hydrolase like |
| chr14_+_91114364 | 0.06 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase |
| chr5_+_132673983 | 0.06 |
ENST00000622422.1
ENST00000231449.7 ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr17_-_7394514 | 0.06 |
ENST00000571802.1
ENST00000619711.5 ENST00000576201.5 ENST00000573213.1 ENST00000324822.15 |
PLSCR3
|
phospholipid scramblase 3 |
| chr10_+_102503985 | 0.06 |
ENST00000369899.6
ENST00000423559.2 |
SUFU
|
SUFU negative regulator of hedgehog signaling |
| chr1_-_37947010 | 0.06 |
ENST00000458109.6
ENST00000373024.8 ENST00000373023.6 |
INPP5B
|
inositol polyphosphate-5-phosphatase B |
| chr1_-_11805949 | 0.06 |
ENST00000376590.9
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chrX_-_53422644 | 0.06 |
ENST00000322213.9
ENST00000375340.10 ENST00000674590.1 |
SMC1A
|
structural maintenance of chromosomes 1A |
| chr19_-_42217667 | 0.06 |
ENST00000336034.8
ENST00000596251.6 ENST00000598200.1 ENST00000598727.5 |
DEDD2
|
death effector domain containing 2 |
| chr19_-_42427379 | 0.06 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr2_+_176107272 | 0.06 |
ENST00000249504.7
|
HOXD11
|
homeobox D11 |
| chr4_-_101347327 | 0.06 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.7 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.2 | 0.5 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 1.6 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.3 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.4 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:0021718 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.2 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.3 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.6 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) gamma-delta T cell differentiation(GO:0042492) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.2 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.4 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0031296 | T-helper 1 cell lineage commitment(GO:0002296) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.0 | GO:0030719 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.0 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 1.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |