Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
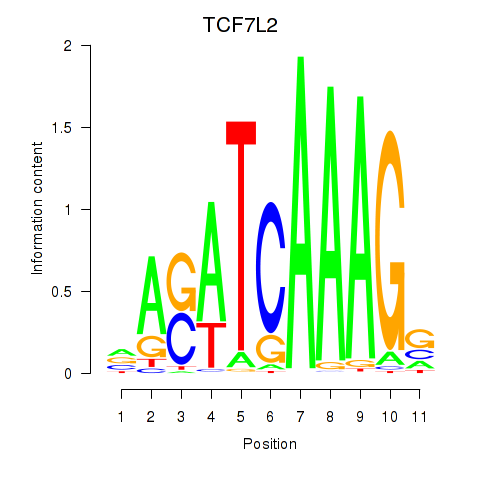
Results for TCF7L2
Z-value: 0.84
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.17 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg38_v1_chr10_+_112950240_112950272 | -0.57 | 1.4e-01 | Click! |
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23570370 | 1.31 |
ENST00000403372.6
ENST00000248484.9 |
TNFRSF19
|
TNF receptor superfamily member 19 |
| chr13_+_101489940 | 1.16 |
ENST00000376162.7
|
ITGBL1
|
integrin subunit beta like 1 |
| chr13_+_75760362 | 0.93 |
ENST00000534657.5
|
LMO7
|
LIM domain 7 |
| chrX_+_153494970 | 0.92 |
ENST00000331595.9
ENST00000431891.1 |
BGN
|
biglycan |
| chr10_+_69088096 | 0.84 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr7_+_94394886 | 0.65 |
ENST00000297268.11
ENST00000620463.1 |
COL1A2
|
collagen type I alpha 2 chain |
| chr13_+_75760659 | 0.63 |
ENST00000526202.5
ENST00000465261.6 |
LMO7
|
LIM domain 7 |
| chr7_+_120988683 | 0.55 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr13_+_75760431 | 0.55 |
ENST00000321797.12
|
LMO7
|
LIM domain 7 |
| chr12_+_1629197 | 0.55 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B |
| chr17_-_69141878 | 0.43 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr1_+_222928415 | 0.43 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr3_-_15797930 | 0.42 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr16_-_73048104 | 0.36 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr4_-_156970903 | 0.35 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr6_-_34248989 | 0.33 |
ENST00000481533.5
ENST00000468145.1 ENST00000476320.6 ENST00000394990.8 |
SMIM29
|
small integral membrane protein 29 |
| chr1_+_60865259 | 0.33 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr4_-_165279679 | 0.32 |
ENST00000505354.2
|
GK3P
|
glycerol kinase 3 pseudogene |
| chr11_-_70717994 | 0.31 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr2_+_200440649 | 0.29 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr3_+_29281552 | 0.28 |
ENST00000452462.5
ENST00000456853.1 |
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr5_-_16742221 | 0.28 |
ENST00000505695.5
|
MYO10
|
myosin X |
| chr8_+_38787218 | 0.28 |
ENST00000317827.9
ENST00000276520.12 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr13_+_31945826 | 0.27 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr3_+_141368497 | 0.27 |
ENST00000321464.7
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_-_153550083 | 0.26 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chrX_+_54809060 | 0.26 |
ENST00000396224.1
|
MAGED2
|
MAGE family member D2 |
| chr11_+_69641146 | 0.26 |
ENST00000227507.3
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr4_+_112860912 | 0.25 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112861053 | 0.25 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112860981 | 0.25 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr19_+_1266653 | 0.24 |
ENST00000586472.5
ENST00000589266.5 |
CIRBP
|
cold inducible RNA binding protein |
| chrX_-_63785510 | 0.24 |
ENST00000437457.6
ENST00000374878.5 ENST00000623517.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr17_+_77451244 | 0.24 |
ENST00000591088.5
|
SEPTIN9
|
septin 9 |
| chr16_+_53208438 | 0.24 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr2_-_55419821 | 0.24 |
ENST00000644630.1
ENST00000471947.2 ENST00000436346.7 ENST00000642200.1 ENST00000413716.7 ENST00000263630.13 ENST00000645072.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr2_-_55419565 | 0.23 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr7_-_139777986 | 0.23 |
ENST00000406875.8
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr15_+_92904447 | 0.22 |
ENST00000626782.2
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr5_+_61332236 | 0.22 |
ENST00000252744.6
|
ZSWIM6
|
zinc finger SWIM-type containing 6 |
| chr1_+_77918128 | 0.22 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chrX_-_100928903 | 0.22 |
ENST00000372956.3
|
XKRX
|
XK related X-linked |
| chr14_-_52791597 | 0.21 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr6_-_34249102 | 0.20 |
ENST00000636500.1
|
SMIM29
|
small integral membrane protein 29 |
| chr5_-_59039454 | 0.20 |
ENST00000358923.10
|
PDE4D
|
phosphodiesterase 4D |
| chr6_+_155216637 | 0.20 |
ENST00000275246.11
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr11_-_111879154 | 0.20 |
ENST00000260257.9
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chrX_-_40097403 | 0.19 |
ENST00000397354.7
|
BCOR
|
BCL6 corepressor |
| chr15_-_89751292 | 0.19 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior bHLH transcription factor 1 |
| chr1_+_206865620 | 0.18 |
ENST00000367098.6
|
IL20
|
interleukin 20 |
| chr19_-_43504711 | 0.18 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology like domain family B member 3 |
| chr17_-_65561640 | 0.17 |
ENST00000618960.4
ENST00000307078.10 |
AXIN2
|
axin 2 |
| chr18_+_6729698 | 0.17 |
ENST00000383472.9
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr5_+_102865805 | 0.16 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr11_-_62707413 | 0.16 |
ENST00000360796.10
ENST00000449636.6 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr10_+_52314272 | 0.16 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr2_+_69013379 | 0.16 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr5_+_112738331 | 0.16 |
ENST00000512211.6
|
APC
|
APC regulator of WNT signaling pathway |
| chr3_+_141387801 | 0.16 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr11_-_62707581 | 0.15 |
ENST00000684475.1
ENST00000683296.1 ENST00000684067.1 ENST00000682223.1 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr20_-_675793 | 0.15 |
ENST00000488788.2
ENST00000246104.7 |
ENSG00000270299.1
SCRT2
|
novel protein scratch family transcriptional repressor 2 |
| chr12_-_49189053 | 0.14 |
ENST00000550767.6
ENST00000546918.1 ENST00000679733.1 ENST00000552924.2 ENST00000301071.12 |
TUBA1A
|
tubulin alpha 1a |
| chr5_-_135954962 | 0.14 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr5_-_157575767 | 0.14 |
ENST00000257527.9
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr17_+_1771688 | 0.14 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr9_-_15510289 | 0.13 |
ENST00000397519.6
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr7_-_80512041 | 0.13 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3 |
| chr17_+_44846318 | 0.13 |
ENST00000591513.5
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr17_-_39864304 | 0.12 |
ENST00000346872.8
|
IKZF3
|
IKAROS family zinc finger 3 |
| chr2_+_203936755 | 0.12 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr2_+_69013170 | 0.12 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr2_-_157628852 | 0.12 |
ENST00000243349.13
|
ACVR1C
|
activin A receptor type 1C |
| chr11_+_114257800 | 0.12 |
ENST00000535401.5
|
NNMT
|
nicotinamide N-methyltransferase |
| chr7_-_151814636 | 0.12 |
ENST00000652047.1
ENST00000650858.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr4_-_68349981 | 0.12 |
ENST00000510746.1
ENST00000355665.7 ENST00000344157.9 |
YTHDC1
|
YTH domain containing 1 |
| chr15_-_89221558 | 0.12 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr19_-_51082883 | 0.12 |
ENST00000650543.2
|
KLK14
|
kallikrein related peptidase 14 |
| chr16_+_29808051 | 0.11 |
ENST00000568544.5
ENST00000569978.1 |
MAZ
|
MYC associated zinc finger protein |
| chr7_-_151814668 | 0.11 |
ENST00000651764.1
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr4_+_2818155 | 0.11 |
ENST00000511747.6
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr5_+_141417659 | 0.11 |
ENST00000398594.4
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr5_+_140107777 | 0.11 |
ENST00000505703.2
ENST00000651386.1 |
PURA
|
purine rich element binding protein A |
| chr11_-_6006946 | 0.10 |
ENST00000641156.1
ENST00000641835.1 |
OR56A4
|
olfactory receptor family 56 subfamily A member 4 |
| chr2_+_203867943 | 0.10 |
ENST00000295854.10
ENST00000487393.1 ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chrX_-_43973382 | 0.10 |
ENST00000642620.1
ENST00000647044.1 |
NDP
|
norrin cystine knot growth factor NDP |
| chr14_+_74348440 | 0.10 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr2_-_71130214 | 0.10 |
ENST00000494660.6
ENST00000244217.6 ENST00000486135.1 |
MCEE
|
methylmalonyl-CoA epimerase |
| chr6_-_166167832 | 0.10 |
ENST00000366876.7
|
TBXT
|
T-box transcription factor T |
| chr12_+_121888751 | 0.09 |
ENST00000261817.6
ENST00000538613.5 ENST00000542602.1 ENST00000541212.6 |
PSMD9
|
proteasome 26S subunit, non-ATPase 9 |
| chr17_+_44004604 | 0.09 |
ENST00000293404.8
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr7_-_148884266 | 0.09 |
ENST00000483967.5
ENST00000320356.7 |
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr7_+_27242796 | 0.09 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1 |
| chr12_-_12338674 | 0.09 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr2_+_69013282 | 0.09 |
ENST00000409829.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr1_-_19923617 | 0.09 |
ENST00000375116.3
|
PLA2G2E
|
phospholipase A2 group IIE |
| chr10_+_99782628 | 0.09 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr16_+_29808125 | 0.09 |
ENST00000568282.1
|
MAZ
|
MYC associated zinc finger protein |
| chr19_-_58519554 | 0.09 |
ENST00000354590.7
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr12_-_49188811 | 0.09 |
ENST00000295766.9
|
TUBA1A
|
tubulin alpha 1a |
| chr17_-_39864140 | 0.09 |
ENST00000623724.3
ENST00000439167.6 ENST00000377945.7 ENST00000394189.6 ENST00000377944.7 ENST00000377958.6 ENST00000535189.5 ENST00000377952.6 |
IKZF3
|
IKAROS family zinc finger 3 |
| chr19_-_18606779 | 0.09 |
ENST00000684169.1
ENST00000392386.8 |
CRLF1
|
cytokine receptor like factor 1 |
| chr2_+_69013414 | 0.09 |
ENST00000681816.1
ENST00000482235.2 |
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr14_-_80959484 | 0.09 |
ENST00000555529.5
ENST00000556042.5 ENST00000556981.5 |
CEP128
|
centrosomal protein 128 |
| chr15_-_55588337 | 0.09 |
ENST00000563719.4
|
PYGO1
|
pygopus family PHD finger 1 |
| chr7_+_27242700 | 0.09 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1 |
| chr19_-_58519751 | 0.09 |
ENST00000600990.1
ENST00000594051.6 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr6_+_63571702 | 0.09 |
ENST00000672924.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr10_+_113553039 | 0.09 |
ENST00000351270.4
|
HABP2
|
hyaluronan binding protein 2 |
| chrX_+_47078380 | 0.09 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chr6_-_89217339 | 0.09 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr10_-_73591330 | 0.08 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr7_-_148884159 | 0.08 |
ENST00000478654.5
ENST00000460911.5 ENST00000350995.6 |
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr10_-_102432565 | 0.08 |
ENST00000369937.5
|
CUEDC2
|
CUE domain containing 2 |
| chr5_+_55160161 | 0.08 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr8_-_143973520 | 0.08 |
ENST00000356346.7
|
PLEC
|
plectin |
| chr1_+_147541491 | 0.07 |
ENST00000683836.1
ENST00000234739.8 |
BCL9
|
BCL9 transcription coactivator |
| chr14_+_32329256 | 0.07 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chrX_+_47078330 | 0.07 |
ENST00000457380.5
|
RGN
|
regucalcin |
| chrX_+_47078434 | 0.07 |
ENST00000397180.6
|
RGN
|
regucalcin |
| chr12_+_8914525 | 0.07 |
ENST00000543824.5
|
PHC1
|
polyhomeotic homolog 1 |
| chr17_+_81712236 | 0.07 |
ENST00000545862.5
ENST00000350690.10 ENST00000331531.9 |
SLC25A10
|
solute carrier family 25 member 10 |
| chr14_+_74763308 | 0.06 |
ENST00000325680.12
ENST00000552421.5 |
YLPM1
|
YLP motif containing 1 |
| chr17_-_42018488 | 0.06 |
ENST00000589773.5
ENST00000674214.1 |
DNAJC7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr4_+_169660062 | 0.06 |
ENST00000507875.5
ENST00000613795.4 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr1_-_201127184 | 0.06 |
ENST00000449188.3
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr17_-_49230728 | 0.06 |
ENST00000503902.1
ENST00000512250.1 ENST00000413580.5 ENST00000511066.5 |
PHOSPHO1
|
phosphoethanolamine/phosphocholine phosphatase 1 |
| chr17_-_75393741 | 0.06 |
ENST00000578961.5
ENST00000392564.5 |
GRB2
|
growth factor receptor bound protein 2 |
| chr17_-_49230770 | 0.06 |
ENST00000310544.9
|
PHOSPHO1
|
phosphoethanolamine/phosphocholine phosphatase 1 |
| chr2_-_88128049 | 0.06 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chr2_+_227813834 | 0.06 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr3_-_177196451 | 0.06 |
ENST00000430069.5
ENST00000630796.2 ENST00000428970.5 |
TBL1XR1
|
TBL1X receptor 1 |
| chr17_-_75393656 | 0.05 |
ENST00000392563.5
|
GRB2
|
growth factor receptor bound protein 2 |
| chr14_+_21797272 | 0.05 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr3_+_137764296 | 0.05 |
ENST00000306087.3
|
SOX14
|
SRY-box transcription factor 14 |
| chr19_-_40285277 | 0.05 |
ENST00000579047.5
ENST00000392038.7 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr4_+_85604146 | 0.05 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_-_134374439 | 0.05 |
ENST00000513145.1
ENST00000249883.10 ENST00000422605.6 |
AMOTL2
|
angiomotin like 2 |
| chr6_+_151809105 | 0.05 |
ENST00000427531.6
|
ESR1
|
estrogen receptor 1 |
| chr5_+_161850597 | 0.05 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr3_-_42410558 | 0.05 |
ENST00000441172.1
ENST00000287748.8 |
LYZL4
|
lysozyme like 4 |
| chr9_-_97944832 | 0.05 |
ENST00000259456.7
|
HEMGN
|
hemogen |
| chr2_-_9630491 | 0.04 |
ENST00000381844.8
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr8_-_142879820 | 0.04 |
ENST00000377675.3
ENST00000517471.5 ENST00000292427.10 |
CYP11B1
|
cytochrome P450 family 11 subfamily B member 1 |
| chr20_-_57265738 | 0.04 |
ENST00000433911.1
|
BMP7
|
bone morphogenetic protein 7 |
| chr10_+_103245887 | 0.04 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1 |
| chr1_+_154272589 | 0.04 |
ENST00000457918.6
ENST00000483970.6 ENST00000328703.12 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr17_-_41065879 | 0.04 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr15_-_93089192 | 0.04 |
ENST00000329082.11
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr1_-_162023826 | 0.04 |
ENST00000294794.8
|
OLFML2B
|
olfactomedin like 2B |
| chr1_-_44141631 | 0.04 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr16_+_72054477 | 0.04 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr11_-_40294089 | 0.04 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr4_-_138242325 | 0.04 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr22_+_40951364 | 0.03 |
ENST00000216225.9
|
RBX1
|
ring-box 1 |
| chr7_+_80646305 | 0.03 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chrX_+_2752024 | 0.03 |
ENST00000644266.2
ENST00000419513.7 ENST00000509484.3 ENST00000381174.10 |
XG
|
Xg glycoprotein (Xg blood group) |
| chr3_-_45915698 | 0.03 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr16_+_72063226 | 0.03 |
ENST00000540303.7
ENST00000356967.6 ENST00000561690.1 |
HPR
|
haptoglobin-related protein |
| chr13_-_71867192 | 0.03 |
ENST00000611519.4
ENST00000620444.4 ENST00000613252.5 |
DACH1
|
dachshund family transcription factor 1 |
| chr11_-_62805429 | 0.03 |
ENST00000294172.7
ENST00000531131.1 ENST00000530875.5 ENST00000531709.6 |
NXF1
|
nuclear RNA export factor 1 |
| chr14_-_63318933 | 0.03 |
ENST00000621500.2
|
GPHB5
|
glycoprotein hormone subunit beta 5 |
| chr11_+_111878926 | 0.03 |
ENST00000528125.5
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr2_-_9630946 | 0.03 |
ENST00000446619.1
ENST00000238081.8 |
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr14_-_93976550 | 0.03 |
ENST00000555019.6
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr6_-_25930678 | 0.03 |
ENST00000377850.8
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr6_-_28252246 | 0.03 |
ENST00000377294.3
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr19_-_39433703 | 0.02 |
ENST00000602153.5
|
RPS16
|
ribosomal protein S16 |
| chr2_+_233712905 | 0.02 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr17_-_47957824 | 0.02 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chr3_+_174859315 | 0.02 |
ENST00000454872.6
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase like 2 |
| chr6_-_25930611 | 0.02 |
ENST00000360488.7
|
SLC17A2
|
solute carrier family 17 member 2 |
| chrX_-_123733023 | 0.02 |
ENST00000245838.13
ENST00000355725.8 |
THOC2
|
THO complex 2 |
| chr3_-_179974254 | 0.02 |
ENST00000468741.5
|
PEX5L
|
peroxisomal biogenesis factor 5 like |
| chr2_+_71130586 | 0.02 |
ENST00000498451.2
ENST00000244230.7 |
MPHOSPH10
|
M-phase phosphoprotein 10 |
| chr11_+_112074287 | 0.02 |
ENST00000532163.5
ENST00000280352.13 ENST00000393047.8 ENST00000526879.5 ENST00000525785.5 |
NKAPD1
|
NKAP domain containing 1 |
| chr8_-_132111159 | 0.02 |
ENST00000673615.1
ENST00000434736.6 |
HHLA1
|
HERV-H LTR-associating 1 |
| chr19_-_40285395 | 0.02 |
ENST00000424901.5
ENST00000578123.5 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr7_+_100015572 | 0.02 |
ENST00000535170.5
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr1_+_109548567 | 0.02 |
ENST00000369851.7
|
GNAI3
|
G protein subunit alpha i3 |
| chr15_+_57219411 | 0.02 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr2_+_112911159 | 0.02 |
ENST00000263326.8
|
IL37
|
interleukin 37 |
| chr9_-_74887720 | 0.02 |
ENST00000449912.6
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6 |
| chr22_+_19962537 | 0.02 |
ENST00000449653.5
|
COMT
|
catechol-O-methyltransferase |
| chr15_+_64460728 | 0.01 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chrX_+_106920393 | 0.01 |
ENST00000336803.2
|
CLDN2
|
claudin 2 |
| chr5_-_24644968 | 0.01 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr9_-_74887914 | 0.01 |
ENST00000360774.6
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6 |
| chrX_+_123860262 | 0.01 |
ENST00000355640.3
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr16_+_50548387 | 0.01 |
ENST00000268459.6
|
NKD1
|
NKD inhibitor of WNT signaling pathway 1 |
| chr2_-_232776555 | 0.01 |
ENST00000438786.1
ENST00000233826.4 ENST00000409779.1 |
KCNJ13
|
potassium inwardly rectifying channel subfamily J member 13 |
| chr7_+_100015588 | 0.01 |
ENST00000324306.11
ENST00000426572.5 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr12_+_8914464 | 0.01 |
ENST00000544916.6
|
PHC1
|
polyhomeotic homolog 1 |
| chr3_+_156120572 | 0.01 |
ENST00000389636.9
ENST00000490337.6 |
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr18_+_32190015 | 0.01 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta |
| chr3_-_33645433 | 0.01 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr7_-_27180230 | 0.01 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr4_-_68349750 | 0.01 |
ENST00000579690.5
|
YTHDC1
|
YTH domain containing 1 |
| chr7_+_80646436 | 0.01 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr5_+_171419635 | 0.01 |
ENST00000274625.6
|
FGF18
|
fibroblast growth factor 18 |
| chr2_+_203867764 | 0.01 |
ENST00000648405.2
|
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.4 | GO:0042662 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of mesodermal cell fate specification(GO:0042662) |
| 0.1 | 0.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.2 | GO:1903627 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.7 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0070221 | succinate transport(GO:0015744) sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.0 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.2 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |