Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
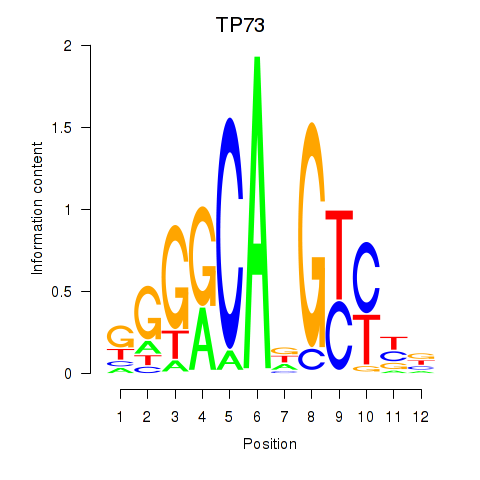
Results for TP73
Z-value: 0.27
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.15 | tumor protein p73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP73 | hg38_v1_chr1_+_3698027_3698058 | 0.67 | 7.1e-02 | Click! |
Activity profile of TP73 motif
Sorted Z-values of TP73 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153615858 | 0.18 |
ENST00000476873.5
|
S100A14
|
S100 calcium binding protein A14 |
| chr19_+_44777860 | 0.17 |
ENST00000341505.4
ENST00000647358.2 |
CBLC
|
Cbl proto-oncogene C |
| chr14_+_75279637 | 0.15 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr19_+_35106510 | 0.13 |
ENST00000648240.1
|
ENSG00000285526.1
|
novel protein |
| chr14_+_75278820 | 0.13 |
ENST00000554617.1
ENST00000554212.5 ENST00000535987.5 ENST00000303562.9 ENST00000555242.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr10_+_46375645 | 0.13 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1 |
| chr10_+_46375619 | 0.12 |
ENST00000584982.7
ENST00000613703.4 |
ANXA8L1
|
annexin A8 like 1 |
| chr17_+_75754618 | 0.11 |
ENST00000584939.1
|
ITGB4
|
integrin subunit beta 4 |
| chr15_+_40844018 | 0.11 |
ENST00000344051.8
ENST00000562057.6 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr3_+_136957948 | 0.10 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr11_+_60429595 | 0.10 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane spanning 4-domains A5 |
| chr1_-_204190324 | 0.09 |
ENST00000638118.1
|
REN
|
renin |
| chr3_-_69386079 | 0.09 |
ENST00000398540.8
|
FRMD4B
|
FERM domain containing 4B |
| chr12_-_56488153 | 0.08 |
ENST00000311966.9
|
GLS2
|
glutaminase 2 |
| chr19_-_291132 | 0.08 |
ENST00000327790.7
|
PLPP2
|
phospholipid phosphatase 2 |
| chr6_+_36676455 | 0.07 |
ENST00000615513.4
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr6_+_36676489 | 0.07 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr11_+_60429567 | 0.07 |
ENST00000300190.7
|
MS4A5
|
membrane spanning 4-domains A5 |
| chr12_-_6635938 | 0.07 |
ENST00000329858.9
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr3_+_50205254 | 0.07 |
ENST00000614032.5
ENST00000445096.5 |
SLC38A3
|
solute carrier family 38 member 3 |
| chr2_+_95274439 | 0.07 |
ENST00000317620.14
ENST00000403131.6 ENST00000317668.8 |
PROM2
|
prominin 2 |
| chr10_-_103452356 | 0.06 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr10_+_46375721 | 0.06 |
ENST00000616785.1
ENST00000611655.1 ENST00000619162.5 |
ENSG00000285402.1
ANXA8L1
|
novel transcript annexin A8 like 1 |
| chr10_-_103452384 | 0.06 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr9_+_35673917 | 0.06 |
ENST00000617161.1
ENST00000378357.9 |
CA9
|
carbonic anhydrase 9 |
| chr3_-_185821092 | 0.05 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr18_-_55586092 | 0.05 |
ENST00000563888.6
ENST00000540999.5 ENST00000627685.2 |
TCF4
|
transcription factor 4 |
| chr18_-_55585773 | 0.05 |
ENST00000563824.5
ENST00000626425.2 ENST00000566514.5 ENST00000568673.5 ENST00000562847.5 ENST00000568147.5 |
TCF4
|
transcription factor 4 |
| chr17_-_44123628 | 0.05 |
ENST00000587135.1
ENST00000225983.10 ENST00000682912.1 ENST00000588703.5 |
HDAC5
|
histone deacetylase 5 |
| chr6_-_46735351 | 0.04 |
ENST00000274793.12
|
PLA2G7
|
phospholipase A2 group VII |
| chr1_+_64745089 | 0.04 |
ENST00000294428.7
ENST00000371072.8 |
RAVER2
|
ribonucleoprotein, PTB binding 2 |
| chr3_+_44976236 | 0.04 |
ENST00000265564.8
|
EXOSC7
|
exosome component 7 |
| chr2_+_74002685 | 0.04 |
ENST00000305799.8
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr12_+_49295138 | 0.04 |
ENST00000257860.9
|
PRPH
|
peripherin |
| chr20_-_54173516 | 0.03 |
ENST00000395955.7
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr12_-_31729010 | 0.03 |
ENST00000537562.5
ENST00000537960.5 ENST00000281471.11 ENST00000536761.5 ENST00000542781.5 ENST00000457428.6 |
AMN1
|
antagonist of mitotic exit network 1 homolog |
| chr4_+_38664189 | 0.03 |
ENST00000514033.1
ENST00000261438.10 |
KLF3
|
Kruppel like factor 3 |
| chr2_+_176116768 | 0.03 |
ENST00000249501.5
|
HOXD10
|
homeobox D10 |
| chr19_+_43533384 | 0.03 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr17_+_39200507 | 0.03 |
ENST00000678573.1
|
RPL19
|
ribosomal protein L19 |
| chr6_-_46735693 | 0.03 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2 group VII |
| chr19_-_35490456 | 0.03 |
ENST00000338897.4
ENST00000484218.6 |
KRTDAP
|
keratinocyte differentiation associated protein |
| chr16_+_30923565 | 0.03 |
ENST00000338343.10
|
FBXL19
|
F-box and leucine rich repeat protein 19 |
| chr16_+_2019777 | 0.03 |
ENST00000566435.4
|
NPW
|
neuropeptide W |
| chrX_+_153642473 | 0.03 |
ENST00000370167.8
|
DUSP9
|
dual specificity phosphatase 9 |
| chr17_+_39200334 | 0.03 |
ENST00000579260.5
ENST00000582193.5 |
RPL19
|
ribosomal protein L19 |
| chr14_-_100569780 | 0.03 |
ENST00000355173.7
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr18_+_13277351 | 0.03 |
ENST00000679091.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr17_+_39200275 | 0.03 |
ENST00000225430.9
|
RPL19
|
ribosomal protein L19 |
| chr11_-_236821 | 0.03 |
ENST00000529382.5
ENST00000528469.1 |
SIRT3
|
sirtuin 3 |
| chr14_+_22086401 | 0.03 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr16_+_56191476 | 0.03 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr19_+_19516216 | 0.03 |
ENST00000507754.9
ENST00000503283.5 ENST00000428459.6 ENST00000555938.1 |
NDUFA13
ENSG00000258674.5
|
NADH:ubiquinone oxidoreductase subunit A13 novel protein |
| chr2_+_219572304 | 0.03 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha |
| chr17_+_39200302 | 0.02 |
ENST00000579374.5
|
RPL19
|
ribosomal protein L19 |
| chr17_-_40867200 | 0.02 |
ENST00000647902.1
ENST00000251643.5 |
KRT12
|
keratin 12 |
| chr16_+_74999003 | 0.02 |
ENST00000335325.9
ENST00000320619.10 |
ZNRF1
|
zinc and ring finger 1 |
| chr17_+_21288029 | 0.02 |
ENST00000526076.6
ENST00000361818.9 ENST00000316920.10 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr17_+_7888783 | 0.02 |
ENST00000330494.12
ENST00000358181.8 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr19_+_41860236 | 0.02 |
ENST00000601492.5
ENST00000600467.6 ENST00000598742.6 ENST00000221975.6 ENST00000598261.2 |
RPS19
|
ribosomal protein S19 |
| chr2_-_26641363 | 0.02 |
ENST00000288861.5
|
CIB4
|
calcium and integrin binding family member 4 |
| chr16_-_8797624 | 0.02 |
ENST00000333050.7
|
TMEM186
|
transmembrane protein 186 |
| chr9_+_36169382 | 0.02 |
ENST00000335119.4
|
CCIN
|
calicin |
| chr19_-_38735405 | 0.02 |
ENST00000597987.5
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chr17_-_76027296 | 0.02 |
ENST00000301607.8
|
EVPL
|
envoplakin |
| chr11_+_66466745 | 0.02 |
ENST00000349459.10
ENST00000320740.12 ENST00000524466.5 ENST00000526296.5 |
PELI3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr4_+_6716131 | 0.02 |
ENST00000320776.5
|
BLOC1S4
|
biogenesis of lysosomal organelles complex 1 subunit 4 |
| chr14_+_103385450 | 0.02 |
ENST00000416682.6
|
MARK3
|
microtubule affinity regulating kinase 3 |
| chr22_-_37580075 | 0.02 |
ENST00000215886.6
|
LGALS2
|
galectin 2 |
| chr8_+_119873710 | 0.02 |
ENST00000523492.5
ENST00000286234.6 |
DEPTOR
|
DEP domain containing MTOR interacting protein |
| chr12_+_82358496 | 0.02 |
ENST00000248306.8
ENST00000548200.5 |
METTL25
|
methyltransferase like 25 |
| chr2_+_158968608 | 0.02 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr16_-_636270 | 0.01 |
ENST00000397665.6
ENST00000397666.6 ENST00000301686.13 ENST00000338401.8 ENST00000397664.8 ENST00000568830.1 ENST00000614890.4 |
METTL26
|
methyltransferase like 26 |
| chr4_-_20984011 | 0.01 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr19_+_41350911 | 0.01 |
ENST00000539627.5
|
TMEM91
|
transmembrane protein 91 |
| chr2_+_89884740 | 0.01 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr10_-_103479240 | 0.01 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr14_+_103385506 | 0.01 |
ENST00000303622.13
|
MARK3
|
microtubule affinity regulating kinase 3 |
| chr10_+_68901258 | 0.01 |
ENST00000373585.8
|
DDX50
|
DExD-box helicase 50 |
| chr20_-_63572455 | 0.01 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2 |
| chr20_-_59940289 | 0.01 |
ENST00000370996.5
|
PPP1R3D
|
protein phosphatase 1 regulatory subunit 3D |
| chr19_+_43716095 | 0.01 |
ENST00000596627.1
|
IRGC
|
immunity related GTPase cinema |
| chr1_+_26159071 | 0.01 |
ENST00000374268.5
|
FAM110D
|
family with sequence similarity 110 member D |
| chr14_+_103385374 | 0.01 |
ENST00000678179.1
ENST00000676938.1 ENST00000678619.1 ENST00000440884.7 ENST00000560417.6 ENST00000679330.1 ENST00000556744.2 ENST00000676897.1 ENST00000677560.1 ENST00000561314.6 ENST00000677829.1 ENST00000677133.1 ENST00000676645.1 ENST00000678175.1 ENST00000429436.7 ENST00000677360.1 ENST00000678237.1 ENST00000677347.1 ENST00000677432.1 |
MARK3
|
microtubule affinity regulating kinase 3 |
| chr7_-_128405930 | 0.01 |
ENST00000470772.5
ENST00000480861.5 ENST00000496200.5 |
IMPDH1
|
inosine monophosphate dehydrogenase 1 |
| chr20_-_54173976 | 0.01 |
ENST00000216862.8
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr10_-_77926724 | 0.01 |
ENST00000372391.7
|
DLG5
|
discs large MAGUK scaffold protein 5 |
| chr1_+_156815633 | 0.01 |
ENST00000392302.7
ENST00000674537.1 |
NTRK1
|
neurotrophic receptor tyrosine kinase 1 |
| chr9_-_135499846 | 0.01 |
ENST00000429260.7
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr11_+_237016 | 0.01 |
ENST00000352303.9
|
PSMD13
|
proteasome 26S subunit, non-ATPase 13 |
| chr17_-_82101850 | 0.01 |
ENST00000583053.1
|
CCDC57
|
coiled-coil domain containing 57 |
| chr17_-_5123102 | 0.01 |
ENST00000250076.7
|
ZNF232
|
zinc finger protein 232 |
| chr2_+_69741974 | 0.01 |
ENST00000409920.5
|
ANXA4
|
annexin A4 |
| chr11_+_236957 | 0.01 |
ENST00000532097.6
ENST00000431206.6 ENST00000528906.5 ENST00000621534.4 |
PSMD13
|
proteasome 26S subunit, non-ATPase 13 |
| chrX_-_132094263 | 0.01 |
ENST00000370879.5
|
FRMD7
|
FERM domain containing 7 |
| chr10_+_62804710 | 0.01 |
ENST00000373783.3
|
ADO
|
2-aminoethanethiol dioxygenase |
| chr5_+_126423122 | 0.01 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr9_+_84669760 | 0.01 |
ENST00000304053.10
|
NTRK2
|
neurotrophic receptor tyrosine kinase 2 |
| chr5_+_126423176 | 0.01 |
ENST00000542322.5
ENST00000544396.5 |
GRAMD2B
|
GRAM domain containing 2B |
| chr16_+_56191728 | 0.01 |
ENST00000638705.1
ENST00000262494.12 |
GNAO1
|
G protein subunit alpha o1 |
| chr16_+_8797813 | 0.01 |
ENST00000268261.9
ENST00000569958.5 |
PMM2
|
phosphomannomutase 2 |
| chr5_+_126423363 | 0.01 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chrX_+_131058340 | 0.01 |
ENST00000276211.10
ENST00000370922.5 |
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr1_+_109539865 | 0.01 |
ENST00000618721.4
ENST00000527748.5 ENST00000616874.1 |
GPR61
|
G protein-coupled receptor 61 |
| chr3_+_48440246 | 0.01 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog |
| chr3_-_48898813 | 0.01 |
ENST00000319017.5
ENST00000430379.5 |
SLC25A20
|
solute carrier family 25 member 20 |
| chr17_-_76027212 | 0.01 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr2_-_27134611 | 0.01 |
ENST00000406567.7
ENST00000260643.7 |
PREB
|
prolactin regulatory element binding |
| chr19_+_12995554 | 0.01 |
ENST00000397661.6
|
NFIX
|
nuclear factor I X |
| chr18_-_4455257 | 0.01 |
ENST00000581527.5
|
DLGAP1
|
DLG associated protein 1 |
| chr3_+_42160160 | 0.01 |
ENST00000341421.7
ENST00000396175.5 |
TRAK1
|
trafficking kinesin protein 1 |
| chr4_+_105710809 | 0.01 |
ENST00000360505.9
ENST00000510865.5 ENST00000509336.5 |
GSTCD
|
glutathione S-transferase C-terminal domain containing |
| chr20_+_59940362 | 0.01 |
ENST00000360816.8
|
FAM217B
|
family with sequence similarity 217 member B |
| chrX_-_139642518 | 0.01 |
ENST00000370573.8
ENST00000338585.6 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr16_+_89918855 | 0.01 |
ENST00000555147.2
|
MC1R
|
melanocortin 1 receptor |
| chr1_-_203086001 | 0.01 |
ENST00000241651.5
|
MYOG
|
myogenin |
| chr19_+_44751251 | 0.00 |
ENST00000444487.1
|
BCL3
|
BCL3 transcription coactivator |
| chr19_+_17215716 | 0.00 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 |
| chr12_-_82358380 | 0.00 |
ENST00000256151.8
ENST00000552377.5 |
CCDC59
|
coiled-coil domain containing 59 |
| chr1_+_16043776 | 0.00 |
ENST00000375679.9
|
CLCNKB
|
chloride voltage-gated channel Kb |
| chr6_+_31547560 | 0.00 |
ENST00000376148.9
ENST00000376145.8 |
NFKBIL1
|
NFKB inhibitor like 1 |
| chr12_-_116881431 | 0.00 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein |
| chr14_-_25050111 | 0.00 |
ENST00000323944.9
|
STXBP6
|
syntaxin binding protein 6 |
| chr1_+_16043736 | 0.00 |
ENST00000619181.4
|
CLCNKB
|
chloride voltage-gated channel Kb |
| chr14_-_25049889 | 0.00 |
ENST00000419632.6
ENST00000396700.5 |
STXBP6
|
syntaxin binding protein 6 |
| chr8_+_144504131 | 0.00 |
ENST00000394955.3
|
GPT
|
glutamic--pyruvic transaminase |
| chr19_+_43716070 | 0.00 |
ENST00000244314.6
|
IRGC
|
immunity related GTPase cinema |
| chr2_-_38602626 | 0.00 |
ENST00000378915.7
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L like |
| chr14_-_24576240 | 0.00 |
ENST00000216336.3
|
CTSG
|
cathepsin G |
| chr19_+_41114430 | 0.00 |
ENST00000331105.7
|
CYP2F1
|
cytochrome P450 family 2 subfamily F member 1 |
| chr2_+_127535746 | 0.00 |
ENST00000428314.5
ENST00000409816.7 |
MYO7B
|
myosin VIIB |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP73
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) cellular response to potassium ion starvation(GO:0051365) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.0 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.0 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |