Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
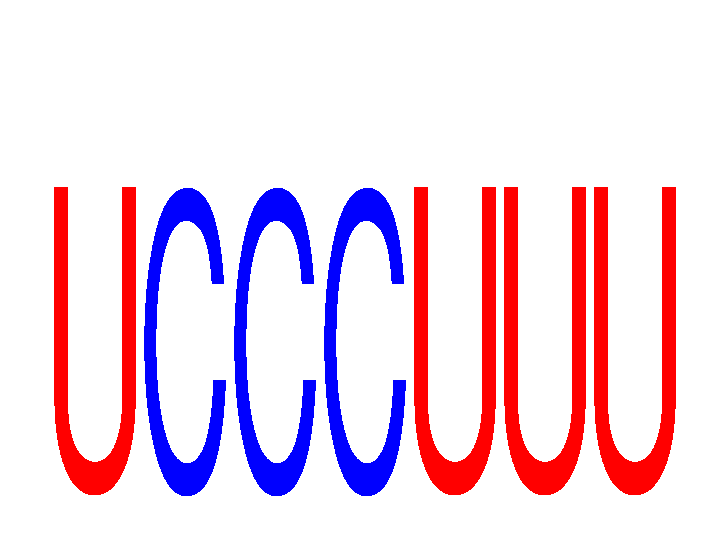
Results for UCCCUUU
Z-value: 0.23
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-204-5p
|
MIMAT0000265 |
|
hsa-miR-211-5p
|
MIMAT0000268 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_95731300 | 0.10 |
ENST00000379982.8
|
RHOBTB3
|
Rho related BTB domain containing 3 |
| chr8_-_92103217 | 0.07 |
ENST00000615601.4
ENST00000523629.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr8_-_121641424 | 0.07 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr10_-_79445617 | 0.06 |
ENST00000372336.4
|
ZCCHC24
|
zinc finger CCHC-type containing 24 |
| chr4_-_65670339 | 0.05 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5 |
| chr9_+_4985227 | 0.05 |
ENST00000381652.4
|
JAK2
|
Janus kinase 2 |
| chr5_-_128538230 | 0.05 |
ENST00000262464.9
|
FBN2
|
fibrillin 2 |
| chr6_+_10555787 | 0.04 |
ENST00000316170.9
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr16_-_73048104 | 0.04 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr3_-_9769910 | 0.04 |
ENST00000256460.8
|
CAMK1
|
calcium/calmodulin dependent protein kinase I |
| chr18_-_28177102 | 0.04 |
ENST00000413878.2
ENST00000269141.8 |
CDH2
|
cadherin 2 |
| chr9_-_20622479 | 0.03 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr6_+_159969070 | 0.03 |
ENST00000356956.6
|
IGF2R
|
insulin like growth factor 2 receptor |
| chr6_-_52577012 | 0.03 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr1_+_63773966 | 0.03 |
ENST00000371079.6
ENST00000371080.5 |
ROR1
|
receptor tyrosine kinase like orphan receptor 1 |
| chr6_+_125790922 | 0.03 |
ENST00000453302.5
ENST00000417494.5 ENST00000392477.7 ENST00000229634.13 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_+_61561561 | 0.03 |
ENST00000474889.6
|
PTPRG
|
protein tyrosine phosphatase receptor type G |
| chr3_-_115071333 | 0.02 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_154405193 | 0.02 |
ENST00000622330.4
ENST00000344086.8 |
IL6R
|
interleukin 6 receptor |
| chr1_+_37793865 | 0.02 |
ENST00000397631.7
|
MANEAL
|
mannosidase endo-alpha like |
| chr8_-_17413345 | 0.02 |
ENST00000180173.10
|
MTMR7
|
myotubularin related protein 7 |
| chr19_-_38224215 | 0.02 |
ENST00000355526.10
ENST00000420980.7 ENST00000614244.4 |
DPF1
|
double PHD fingers 1 |
| chr5_-_147453888 | 0.02 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr16_-_30787169 | 0.02 |
ENST00000262525.6
|
ZNF629
|
zinc finger protein 629 |
| chr3_-_172711005 | 0.02 |
ENST00000424772.2
ENST00000475381.7 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr6_-_158818225 | 0.02 |
ENST00000337147.11
|
EZR
|
ezrin |
| chr4_-_113761927 | 0.02 |
ENST00000296402.9
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr12_+_65824475 | 0.02 |
ENST00000403681.7
|
HMGA2
|
high mobility group AT-hook 2 |
| chr18_-_63319987 | 0.02 |
ENST00000398117.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr2_-_199457931 | 0.02 |
ENST00000417098.6
|
SATB2
|
SATB homeobox 2 |
| chr13_-_49936298 | 0.02 |
ENST00000613924.1
ENST00000361840.8 |
SPRYD7
|
SPRY domain containing 7 |
| chr3_+_132417487 | 0.02 |
ENST00000260818.11
|
DNAJC13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chrX_-_135973975 | 0.02 |
ENST00000305963.3
ENST00000680510.1 ENST00000679621.1 |
MMGT1
|
membrane magnesium transporter 1 |
| chr7_+_138460238 | 0.01 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chrX_+_28587411 | 0.01 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chr1_+_89633086 | 0.01 |
ENST00000370454.9
|
LRRC8C
|
leucine rich repeat containing 8 VRAC subunit C |
| chr5_-_151686908 | 0.01 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr10_-_74150781 | 0.01 |
ENST00000355264.9
ENST00000372745.1 |
AP3M1
|
adaptor related protein complex 3 subunit mu 1 |
| chr5_-_143403611 | 0.01 |
ENST00000394464.7
ENST00000231509.7 |
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr15_+_76336755 | 0.01 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr2_-_68252482 | 0.01 |
ENST00000234310.8
|
PPP3R1
|
protein phosphatase 3 regulatory subunit B, alpha |
| chr16_+_30395400 | 0.01 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr15_-_34336749 | 0.01 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr22_-_28679865 | 0.01 |
ENST00000397906.6
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr2_+_200811882 | 0.01 |
ENST00000409600.6
|
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr17_-_8295342 | 0.01 |
ENST00000579192.5
ENST00000577745.2 |
SLC25A35
|
solute carrier family 25 member 35 |
| chr1_-_38859669 | 0.01 |
ENST00000373001.4
|
RRAGC
|
Ras related GTP binding C |
| chr3_-_48662877 | 0.01 |
ENST00000164024.5
|
CELSR3
|
cadherin EGF LAG seven-pass G-type receptor 3 |
| chr11_+_35618450 | 0.01 |
ENST00000317811.6
|
FJX1
|
four-jointed box kinase 1 |
| chr12_-_108731505 | 0.01 |
ENST00000261401.8
ENST00000552871.5 |
CORO1C
|
coronin 1C |
| chr8_-_90646074 | 0.01 |
ENST00000458549.7
|
TMEM64
|
transmembrane protein 64 |
| chr5_+_172834225 | 0.01 |
ENST00000393784.8
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr20_+_20368096 | 0.01 |
ENST00000310227.3
|
INSM1
|
INSM transcriptional repressor 1 |
| chr13_-_74133892 | 0.01 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12 |
| chr5_-_131796921 | 0.01 |
ENST00000307968.11
ENST00000307954.12 |
FNIP1
|
folliculin interacting protein 1 |
| chr1_-_70205531 | 0.01 |
ENST00000370952.4
|
LRRC40
|
leucine rich repeat containing 40 |
| chr11_+_63938971 | 0.01 |
ENST00000539656.5
ENST00000377793.9 |
NAA40
|
N-alpha-acetyltransferase 40, NatD catalytic subunit |
| chr8_+_43140405 | 0.01 |
ENST00000379644.9
|
HGSNAT
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr3_+_30606574 | 0.01 |
ENST00000295754.10
ENST00000359013.4 |
TGFBR2
|
transforming growth factor beta receptor 2 |
| chr8_-_56211257 | 0.01 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr11_+_58579052 | 0.01 |
ENST00000316059.7
|
ZFP91
|
ZFP91 zinc finger protein, atypical E3 ubiquitin ligase |
| chr3_-_15332526 | 0.01 |
ENST00000383791.8
|
SH3BP5
|
SH3 domain binding protein 5 |
| chr16_+_46884323 | 0.01 |
ENST00000340124.9
|
GPT2
|
glutamic--pyruvic transaminase 2 |
| chr9_-_27573391 | 0.01 |
ENST00000644136.1
ENST00000380003.8 |
C9orf72
|
C9orf72-SMCR8 complex subunit |
| chr12_-_48788995 | 0.01 |
ENST00000550422.5
ENST00000357869.8 |
ADCY6
|
adenylate cyclase 6 |
| chr15_+_38252792 | 0.01 |
ENST00000299084.9
|
SPRED1
|
sprouty related EVH1 domain containing 1 |
| chr12_+_123384078 | 0.01 |
ENST00000402868.8
|
KMT5A
|
lysine methyltransferase 5A |
| chrX_+_71533095 | 0.01 |
ENST00000373719.8
ENST00000373701.7 |
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr12_+_57216779 | 0.01 |
ENST00000349394.6
|
NXPH4
|
neurexophilin 4 |
| chr22_+_40177917 | 0.01 |
ENST00000454349.7
ENST00000335727.13 |
TNRC6B
|
trinucleotide repeat containing adaptor 6B |
| chr16_-_71724700 | 0.01 |
ENST00000568954.5
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chrX_+_10015226 | 0.01 |
ENST00000380861.9
|
WWC3
|
WWC family member 3 |
| chr5_+_173888335 | 0.01 |
ENST00000265085.10
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_+_108719473 | 0.01 |
ENST00000283195.11
|
RANBP2
|
RAN binding protein 2 |
| chr1_+_78004930 | 0.01 |
ENST00000370763.6
|
DNAJB4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr3_-_142448060 | 0.01 |
ENST00000264951.8
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr3_+_152299392 | 0.01 |
ENST00000498502.5
ENST00000545754.5 ENST00000357472.7 ENST00000324196.9 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr4_-_110198650 | 0.01 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr12_+_20368495 | 0.01 |
ENST00000359062.4
|
PDE3A
|
phosphodiesterase 3A |
| chr13_+_34942263 | 0.01 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr6_-_89352706 | 0.01 |
ENST00000435041.3
|
UBE2J1
|
ubiquitin conjugating enzyme E2 J1 |
| chr12_-_38905584 | 0.01 |
ENST00000331366.10
|
CPNE8
|
copine 8 |
| chr17_+_29390326 | 0.01 |
ENST00000261716.8
|
TAOK1
|
TAO kinase 1 |
| chr9_+_110048598 | 0.01 |
ENST00000434623.6
ENST00000374525.5 |
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr3_+_155870623 | 0.01 |
ENST00000295920.7
ENST00000496455.7 |
GMPS
|
guanine monophosphate synthase |
| chr11_-_66347560 | 0.01 |
ENST00000311181.5
|
B4GAT1
|
beta-1,4-glucuronyltransferase 1 |
| chr20_+_31968141 | 0.01 |
ENST00000562532.3
|
XKR7
|
XK related 7 |
| chr5_+_96936071 | 0.01 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase |
| chr1_-_6180265 | 0.01 |
ENST00000262450.8
|
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr2_+_5692357 | 0.01 |
ENST00000322002.5
|
SOX11
|
SRY-box transcription factor 11 |
| chr5_+_65926556 | 0.01 |
ENST00000380943.6
ENST00000416865.6 ENST00000380935.5 ENST00000284037.10 |
ERBIN
|
erbb2 interacting protein |
| chr12_-_57742120 | 0.01 |
ENST00000257897.7
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr7_-_128343823 | 0.01 |
ENST00000415472.6
ENST00000478061.5 ENST00000223073.6 ENST00000459726.1 |
RBM28
|
RNA binding motif protein 28 |
| chr17_-_4263847 | 0.01 |
ENST00000570535.5
ENST00000574367.5 ENST00000341657.9 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chrX_+_49922605 | 0.01 |
ENST00000376088.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr2_+_148644706 | 0.01 |
ENST00000258484.11
|
EPC2
|
enhancer of polycomb homolog 2 |
| chr13_+_113297217 | 0.01 |
ENST00000332556.5
|
LAMP1
|
lysosomal associated membrane protein 1 |
| chr15_+_56918612 | 0.01 |
ENST00000438423.6
ENST00000267811.9 ENST00000333725.10 ENST00000559609.5 |
TCF12
|
transcription factor 12 |
| chr11_+_35662739 | 0.01 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44 |
| chr10_+_91923762 | 0.01 |
ENST00000265990.11
|
BTAF1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr10_+_70815889 | 0.01 |
ENST00000373202.8
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr17_-_68291116 | 0.01 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chr21_-_14383125 | 0.01 |
ENST00000285667.4
|
HSPA13
|
heat shock protein family A (Hsp70) member 13 |
| chr1_+_32465046 | 0.01 |
ENST00000609129.2
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr9_+_109780292 | 0.01 |
ENST00000374530.7
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr17_+_30378903 | 0.01 |
ENST00000225719.9
|
CPD
|
carboxypeptidase D |
| chr9_+_74497308 | 0.00 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr12_+_54008961 | 0.00 |
ENST00000040584.6
|
HOXC8
|
homeobox C8 |
| chr1_-_70354673 | 0.00 |
ENST00000370944.9
ENST00000262346.6 |
ANKRD13C
|
ankyrin repeat domain 13C |
| chr3_+_184155310 | 0.00 |
ENST00000313143.9
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr19_-_474880 | 0.00 |
ENST00000382696.7
ENST00000315489.5 |
ODF3L2
|
outer dense fiber of sperm tails 3 like 2 |
| chr15_-_26773022 | 0.00 |
ENST00000311550.10
ENST00000622697.4 |
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3 |
| chr10_+_86756580 | 0.00 |
ENST00000372037.8
|
BMPR1A
|
bone morphogenetic protein receptor type 1A |
| chr19_+_10420474 | 0.00 |
ENST00000380702.7
|
PDE4A
|
phosphodiesterase 4A |
| chrX_-_132218124 | 0.00 |
ENST00000342983.6
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chrX_-_15854791 | 0.00 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr5_-_180861248 | 0.00 |
ENST00000502412.2
ENST00000512132.5 ENST00000506439.5 |
ZFP62
|
ZFP62 zinc finger protein |
| chr16_+_53703963 | 0.00 |
ENST00000636218.1
ENST00000637001.1 ENST00000471389.6 ENST00000637969.1 ENST00000640179.1 |
FTO
|
FTO alpha-ketoglutarate dependent dioxygenase |
| chr12_+_50925007 | 0.00 |
ENST00000332160.5
|
METTL7A
|
methyltransferase like 7A |
| chr8_+_42896883 | 0.00 |
ENST00000307602.9
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr1_-_161309961 | 0.00 |
ENST00000533357.5
ENST00000672602.2 ENST00000526189.3 |
MPZ
|
myelin protein zero |
| chr11_+_34051722 | 0.00 |
ENST00000341394.9
ENST00000389645.7 |
CAPRIN1
|
cell cycle associated protein 1 |
| chr2_-_229921963 | 0.00 |
ENST00000343290.5
ENST00000389044.8 ENST00000675453.1 ENST00000675903.1 ENST00000283943.9 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_+_11272413 | 0.00 |
ENST00000446450.6
ENST00000354956.9 ENST00000354449.7 ENST00000419112.5 |
ATG7
|
autophagy related 7 |
| chr10_+_58385395 | 0.00 |
ENST00000487519.6
ENST00000373895.7 |
TFAM
|
transcription factor A, mitochondrial |
| chr10_-_119596495 | 0.00 |
ENST00000369093.6
|
TIAL1
|
TIA1 cytotoxic granule associated RNA binding protein like 1 |
| chr10_-_87863533 | 0.00 |
ENST00000445946.5
|
KLLN
|
killin, p53 regulated DNA replication inhibitor |
| chr17_+_63622406 | 0.00 |
ENST00000579585.5
ENST00000361733.8 ENST00000584573.5 ENST00000361357.7 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr18_+_9136757 | 0.00 |
ENST00000262126.9
ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr7_+_107580215 | 0.00 |
ENST00000465919.5
ENST00000005259.9 ENST00000445771.6 ENST00000479917.5 ENST00000421217.5 ENST00000457837.5 |
BCAP29
|
B cell receptor associated protein 29 |
| chr1_-_22143088 | 0.00 |
ENST00000290167.11
|
WNT4
|
Wnt family member 4 |
| chr1_+_2228310 | 0.00 |
ENST00000378536.5
|
SKI
|
SKI proto-oncogene |
| chr2_-_105329685 | 0.00 |
ENST00000393359.7
|
TGFBRAP1
|
transforming growth factor beta receptor associated protein 1 |
| chr1_+_193121950 | 0.00 |
ENST00000367435.5
|
CDC73
|
cell division cycle 73 |
| chr3_-_52278620 | 0.00 |
ENST00000296490.8
|
WDR82
|
WD repeat domain 82 |
| chr6_-_8435480 | 0.00 |
ENST00000379660.4
|
SLC35B3
|
solute carrier family 35 member B3 |
| chr1_+_32013848 | 0.00 |
ENST00000327300.12
ENST00000492989.1 |
KHDRBS1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr2_+_43838963 | 0.00 |
ENST00000272286.4
|
ABCG8
|
ATP binding cassette subfamily G member 8 |
| chr6_-_8064333 | 0.00 |
ENST00000543936.7
ENST00000397457.7 |
BLOC1S5
|
biogenesis of lysosomal organelles complex 1 subunit 5 |
| chr6_+_36442985 | 0.00 |
ENST00000373731.7
ENST00000483557.5 ENST00000498267.5 ENST00000449081.6 ENST00000460983.1 |
KCTD20
|
potassium channel tetramerization domain containing 20 |
| chr4_-_139177185 | 0.00 |
ENST00000394235.6
|
ELF2
|
E74 like ETS transcription factor 2 |
| chr16_-_88941198 | 0.00 |
ENST00000327483.9
ENST00000564416.1 |
CBFA2T3
|
CBFA2/RUNX1 partner transcriptional co-repressor 3 |
| chr13_+_97953652 | 0.00 |
ENST00000460070.6
ENST00000481455.6 ENST00000261574.10 ENST00000651721.2 ENST00000493281.6 ENST00000463157.6 ENST00000471898.5 ENST00000489058.6 ENST00000481689.6 |
IPO5
|
importin 5 |
| chr6_-_43575966 | 0.00 |
ENST00000265351.12
|
XPO5
|
exportin 5 |
| chr20_+_58309704 | 0.00 |
ENST00000244040.4
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr16_+_70114306 | 0.00 |
ENST00000288050.9
ENST00000398122.7 ENST00000568530.5 |
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit |
| chr2_+_148021001 | 0.00 |
ENST00000407073.5
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr4_-_41748713 | 0.00 |
ENST00000226382.4
|
PHOX2B
|
paired like homeobox 2B |
| chr12_-_49060742 | 0.00 |
ENST00000301067.12
ENST00000683543.1 |
KMT2D
|
lysine methyltransferase 2D |
| chr5_-_39074377 | 0.00 |
ENST00000514735.1
ENST00000357387.8 ENST00000296782.9 |
RICTOR
|
RPTOR independent companion of MTOR complex 2 |
| chr14_-_73787244 | 0.00 |
ENST00000394071.6
|
MIDEAS
|
mitotic deacetylase associated SANT domain protein |
| chr3_+_10141763 | 0.00 |
ENST00000256474.3
ENST00000345392.2 |
VHL
|
von Hippel-Lindau tumor suppressor |
| chr10_-_931684 | 0.00 |
ENST00000316157.8
|
LARP4B
|
La ribonucleoprotein 4B |
| chr7_-_32490361 | 0.00 |
ENST00000410044.5
ENST00000450169.7 ENST00000409987.5 ENST00000409782.5 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_+_32142454 | 0.00 |
ENST00000333942.10
ENST00000358365.7 ENST00000545287.7 |
RHOT1
|
ras homolog family member T1 |
| chr4_-_67701113 | 0.00 |
ENST00000420827.2
ENST00000322244.10 |
UBA6
|
ubiquitin like modifier activating enzyme 6 |
| chr5_-_56952107 | 0.00 |
ENST00000381226.7
ENST00000381199.8 ENST00000381213.7 |
MIER3
|
MIER family member 3 |
| chr8_+_103819244 | 0.00 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_81770039 | 0.00 |
ENST00000222390.11
ENST00000453411.6 ENST00000457544.7 ENST00000444829.7 |
HGF
|
hepatocyte growth factor |
| chr12_+_76764109 | 0.00 |
ENST00000426126.7
|
ZDHHC17
|
zinc finger DHHC-type palmitoyltransferase 17 |
| chr12_+_62466791 | 0.00 |
ENST00000641654.1
ENST00000546600.5 ENST00000393630.8 ENST00000552738.5 ENST00000393629.6 ENST00000552115.5 |
MON2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chrX_-_14873027 | 0.00 |
ENST00000452869.1
ENST00000398334.5 ENST00000650831.1 ENST00000324138.7 |
FANCB
|
FA complementation group B |
| chr3_-_120349294 | 0.00 |
ENST00000295628.4
|
LRRC58
|
leucine rich repeat containing 58 |
| chr18_+_34978244 | 0.00 |
ENST00000436190.6
|
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr1_+_21509415 | 0.00 |
ENST00000374840.8
ENST00000539907.5 ENST00000540617.5 |
ALPL
|
alkaline phosphatase, biomineralization associated |
| chr6_+_45328203 | 0.00 |
ENST00000371432.7
ENST00000647337.2 ENST00000371438.5 |
RUNX2
|
RUNX family transcription factor 2 |
| chr4_-_38856807 | 0.00 |
ENST00000506146.5
ENST00000436693.6 ENST00000508254.5 ENST00000514655.1 |
TLR1
TLR6
|
toll like receptor 1 toll like receptor 6 |
| chr3_+_5187697 | 0.00 |
ENST00000256497.9
|
EDEM1
|
ER degradation enhancing alpha-mannosidase like protein 1 |
| chr10_-_59906509 | 0.00 |
ENST00000263102.7
|
CCDC6
|
coiled-coil domain containing 6 |
| chr4_-_23890035 | 0.00 |
ENST00000507380.1
ENST00000264867.7 |
PPARGC1A
|
PPARG coactivator 1 alpha |
| chr7_+_101154445 | 0.00 |
ENST00000337619.11
ENST00000429457.1 |
AP1S1
|
adaptor related protein complex 1 subunit sigma 1 |
| chr3_+_33114007 | 0.00 |
ENST00000320954.11
|
CRTAP
|
cartilage associated protein |
| chr19_-_10010492 | 0.00 |
ENST00000264828.4
|
COL5A3
|
collagen type V alpha 3 chain |
| chr15_+_71096941 | 0.00 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr12_+_122078740 | 0.00 |
ENST00000319080.12
|
MLXIP
|
MLX interacting protein |
| chr13_+_49997019 | 0.00 |
ENST00000420995.6
ENST00000356017.8 ENST00000378182.4 ENST00000457662.2 |
TRIM13
|
tripartite motif containing 13 |
| chr7_+_139231225 | 0.00 |
ENST00000473989.8
|
UBN2
|
ubinuclein 2 |
| chr14_+_57268963 | 0.00 |
ENST00000261558.8
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr2_-_131093378 | 0.00 |
ENST00000409185.5
ENST00000389915.4 |
FAM168B
|
family with sequence similarity 168 member B |
| chr22_+_20917398 | 0.00 |
ENST00000354336.8
|
CRKL
|
CRK like proto-oncogene, adaptor protein |
| chr7_-_140398456 | 0.00 |
ENST00000340308.7
ENST00000447932.6 ENST00000326232.14 ENST00000469193.5 |
SLC37A3
|
solute carrier family 37 member 3 |
| chr2_-_171160833 | 0.00 |
ENST00000360843.7
ENST00000431350.7 |
TLK1
|
tousled like kinase 1 |
| chr7_+_94656325 | 0.00 |
ENST00000482108.1
ENST00000488574.5 ENST00000612748.1 ENST00000613043.1 |
PEG10
|
paternally expressed 10 |
| chr5_-_16936231 | 0.00 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr2_+_191678122 | 0.00 |
ENST00000425611.9
ENST00000410026.7 |
NABP1
|
nucleic acid binding protein 1 |
| chr10_+_12129637 | 0.00 |
ENST00000379051.5
ENST00000379033.7 ENST00000441368.5 ENST00000298428.14 ENST00000304267.12 ENST00000379020.8 ENST00000379017.7 |
SEC61A2
|
SEC61 translocon subunit alpha 2 |
| chr22_+_39456996 | 0.00 |
ENST00000341184.7
|
MGAT3
|
beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase |
| chr7_-_127585566 | 0.00 |
ENST00000321407.3
|
GCC1
|
GRIP and coiled-coil domain containing 1 |
| chr2_+_207529892 | 0.00 |
ENST00000432329.6
ENST00000445803.5 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr15_+_43185382 | 0.00 |
ENST00000300213.9
|
CCNDBP1
|
cyclin D1 binding protein 1 |
| chr11_-_6234618 | 0.00 |
ENST00000449352.7
|
FAM160A2
|
family with sequence similarity 160 member A2 |
| chr9_+_130579556 | 0.00 |
ENST00000319725.10
|
FUBP3
|
far upstream element binding protein 3 |
| chr11_-_27472698 | 0.00 |
ENST00000389858.4
ENST00000379214.9 |
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr18_+_33578213 | 0.00 |
ENST00000681521.1
ENST00000269197.12 |
ASXL3
|
ASXL transcriptional regulator 3 |
| chr15_-_68432151 | 0.00 |
ENST00000423218.6
ENST00000315757.9 |
ITGA11
|
integrin subunit alpha 11 |
| chr15_-_74938027 | 0.00 |
ENST00000564811.1
ENST00000562233.5 ENST00000322347.11 ENST00000567270.5 ENST00000568783.5 |
COX5A
|
cytochrome c oxidase subunit 5A |
| chr14_-_26597430 | 0.00 |
ENST00000344429.9
ENST00000574031.1 ENST00000465357.6 ENST00000547619.5 |
NOVA1
|
NOVA alternative splicing regulator 1 |
| chr10_-_80205551 | 0.00 |
ENST00000372231.7
ENST00000438331.5 |
ANXA11
|
annexin A11 |
| chr7_-_105388881 | 0.00 |
ENST00000460391.5
ENST00000393651.8 |
SRPK2
|
SRSF protein kinase 2 |
| chr8_-_42896677 | 0.00 |
ENST00000526349.5
ENST00000534961.5 |
RNF170
|
ring finger protein 170 |
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.0 | GO:1902728 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |