Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
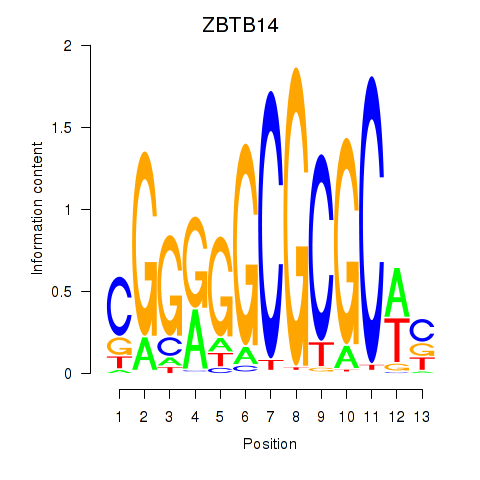
Results for ZBTB14
Z-value: 0.52
Transcription factors associated with ZBTB14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB14
|
ENSG00000198081.11 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB14 | hg38_v1_chr18_-_5295679_5295737, hg38_v1_chr18_-_5296002_5296046 | 0.49 | 2.2e-01 | Click! |
Activity profile of ZBTB14 motif
Sorted Z-values of ZBTB14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_123972824 | 0.28 |
ENST00000238156.8
ENST00000545037.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr12_-_123972709 | 0.27 |
ENST00000545891.5
|
CCDC92
|
coiled-coil domain containing 92 |
| chrX_-_107775951 | 0.24 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr5_+_76403266 | 0.24 |
ENST00000274364.11
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr8_-_141000937 | 0.22 |
ENST00000520892.5
|
PTK2
|
protein tyrosine kinase 2 |
| chr5_+_93584916 | 0.20 |
ENST00000647447.1
ENST00000615873.1 |
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr11_+_114060204 | 0.18 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr8_+_96493803 | 0.18 |
ENST00000518385.5
ENST00000302190.9 |
SDC2
|
syndecan 2 |
| chr2_-_225042433 | 0.17 |
ENST00000258390.12
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr5_+_120464236 | 0.17 |
ENST00000407149.7
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr10_+_21534440 | 0.17 |
ENST00000650772.1
ENST00000652497.1 |
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr4_+_144646145 | 0.17 |
ENST00000296575.8
ENST00000434550.2 |
HHIP
|
hedgehog interacting protein |
| chr2_+_27890716 | 0.17 |
ENST00000344773.6
ENST00000379624.6 ENST00000342045.6 ENST00000379632.6 ENST00000361704.6 |
BABAM2
|
BRISC and BRCA1 A complex member 2 |
| chr9_+_17579059 | 0.17 |
ENST00000380607.5
|
SH3GL2
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr6_+_108559742 | 0.16 |
ENST00000343882.10
|
FOXO3
|
forkhead box O3 |
| chr22_-_31345770 | 0.16 |
ENST00000215919.3
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr10_+_21534315 | 0.16 |
ENST00000377091.7
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr11_+_114059755 | 0.16 |
ENST00000684295.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr7_-_45921264 | 0.16 |
ENST00000613132.5
ENST00000381083.9 ENST00000381086.9 |
IGFBP3
|
insulin like growth factor binding protein 3 |
| chr14_+_73569266 | 0.15 |
ENST00000613168.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr14_+_73569115 | 0.15 |
ENST00000622407.4
ENST00000238651.10 |
ACOT2
|
acyl-CoA thioesterase 2 |
| chr20_+_36461460 | 0.15 |
ENST00000482872.5
ENST00000495241.5 |
DLGAP4
|
DLG associated protein 4 |
| chr11_+_114059702 | 0.15 |
ENST00000335953.9
ENST00000684612.1 ENST00000682810.1 ENST00000544220.1 |
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr5_-_108382080 | 0.14 |
ENST00000542267.7
|
FBXL17
|
F-box and leucine rich repeat protein 17 |
| chr6_-_46491431 | 0.14 |
ENST00000371374.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr5_+_172641241 | 0.14 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr6_-_46491956 | 0.14 |
ENST00000306764.11
|
RCAN2
|
regulator of calcineurin 2 |
| chr3_-_186108501 | 0.13 |
ENST00000422039.1
ENST00000434744.5 |
ETV5
|
ETS variant transcription factor 5 |
| chr14_+_73537346 | 0.13 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chrX_+_9786420 | 0.13 |
ENST00000380913.8
|
SHROOM2
|
shroom family member 2 |
| chr8_-_141001217 | 0.13 |
ENST00000522684.5
ENST00000524357.5 ENST00000521332.5 ENST00000524040.5 ENST00000519881.5 ENST00000520045.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr2_-_27890348 | 0.13 |
ENST00000302188.8
|
RBKS
|
ribokinase |
| chr17_+_46590669 | 0.13 |
ENST00000398238.8
|
NSF
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr15_+_75843438 | 0.12 |
ENST00000267938.9
|
UBE2Q2
|
ubiquitin conjugating enzyme E2 Q2 |
| chr5_+_149141483 | 0.12 |
ENST00000326685.11
ENST00000309868.12 |
ABLIM3
|
actin binding LIM protein family member 3 |
| chr17_+_17782108 | 0.12 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr17_+_75047205 | 0.12 |
ENST00000322444.7
|
KCTD2
|
potassium channel tetramerization domain containing 2 |
| chr14_+_73537135 | 0.12 |
ENST00000311148.9
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr5_+_149141573 | 0.12 |
ENST00000506113.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chrX_-_107775740 | 0.12 |
ENST00000372383.9
|
TSC22D3
|
TSC22 domain family member 3 |
| chr5_+_149141817 | 0.12 |
ENST00000504238.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chr2_+_56183973 | 0.12 |
ENST00000407595.3
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr21_-_44817968 | 0.11 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin like modifier 3 |
| chr14_+_99971442 | 0.11 |
ENST00000402714.6
|
EVL
|
Enah/Vasp-like |
| chr10_+_21534213 | 0.11 |
ENST00000377100.8
ENST00000377072.8 ENST00000307729.12 |
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr17_-_48037760 | 0.11 |
ENST00000621465.5
|
COPZ2
|
COPI coat complex subunit zeta 2 |
| chr17_-_19745369 | 0.11 |
ENST00000573368.5
ENST00000457500.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr16_+_67029359 | 0.11 |
ENST00000565389.1
|
CBFB
|
core-binding factor subunit beta |
| chr15_+_75843307 | 0.11 |
ENST00000569423.5
|
UBE2Q2
|
ubiquitin conjugating enzyme E2 Q2 |
| chr2_-_228181669 | 0.11 |
ENST00000392056.8
|
SPHKAP
|
SPHK1 interactor, AKAP domain containing |
| chr2_-_228181612 | 0.11 |
ENST00000344657.5
|
SPHKAP
|
SPHK1 interactor, AKAP domain containing |
| chr13_+_113584683 | 0.11 |
ENST00000375370.10
|
TFDP1
|
transcription factor Dp-1 |
| chr12_+_53046969 | 0.11 |
ENST00000379902.7
|
TNS2
|
tensin 2 |
| chr1_-_11691471 | 0.10 |
ENST00000376672.5
|
MAD2L2
|
mitotic arrest deficient 2 like 2 |
| chr5_-_19988179 | 0.10 |
ENST00000502796.5
ENST00000382275.6 ENST00000511273.1 |
CDH18
|
cadherin 18 |
| chr5_-_169300782 | 0.10 |
ENST00000332966.8
|
SLIT3
|
slit guidance ligand 3 |
| chr22_-_31346143 | 0.10 |
ENST00000405309.7
ENST00000351933.8 |
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr16_+_1989949 | 0.10 |
ENST00000248121.7
ENST00000618464.1 |
SYNGR3
|
synaptogyrin 3 |
| chr21_-_44818119 | 0.10 |
ENST00000411651.6
|
SUMO3
|
small ubiquitin like modifier 3 |
| chr1_-_1600081 | 0.10 |
ENST00000422725.4
|
FNDC10
|
fibronectin type III domain containing 10 |
| chr6_-_166627244 | 0.10 |
ENST00000265678.9
|
RPS6KA2
|
ribosomal protein S6 kinase A2 |
| chr18_+_6729698 | 0.10 |
ENST00000383472.9
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr6_+_123803853 | 0.10 |
ENST00000368417.6
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr7_-_73557671 | 0.10 |
ENST00000411832.5
ENST00000455335.2 ENST00000223368.7 |
BCL7B
|
BAF chromatin remodeling complex subunit BCL7B |
| chr6_+_84033717 | 0.10 |
ENST00000257776.5
|
MRAP2
|
melanocortin 2 receptor accessory protein 2 |
| chr12_-_31590967 | 0.10 |
ENST00000354285.8
|
DENND5B
|
DENN domain containing 5B |
| chr1_-_11691646 | 0.10 |
ENST00000235310.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2 |
| chr21_-_44818043 | 0.10 |
ENST00000397898.7
ENST00000332859.11 |
SUMO3
|
small ubiquitin like modifier 3 |
| chr14_+_105314711 | 0.10 |
ENST00000447393.6
ENST00000547217.5 |
PACS2
|
phosphofurin acidic cluster sorting protein 2 |
| chr8_-_12755457 | 0.10 |
ENST00000398246.8
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr1_-_11691608 | 0.10 |
ENST00000376667.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2 |
| chr14_-_89619118 | 0.09 |
ENST00000345097.8
ENST00000555855.5 ENST00000555353.5 |
FOXN3
|
forkhead box N3 |
| chr2_-_175005357 | 0.09 |
ENST00000409156.7
ENST00000444573.2 ENST00000409900.9 |
CHN1
|
chimerin 1 |
| chr14_-_60649449 | 0.09 |
ENST00000645694.3
|
SIX1
|
SIX homeobox 1 |
| chr12_-_124567464 | 0.09 |
ENST00000458234.5
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr5_-_111757382 | 0.09 |
ENST00000453526.6
ENST00000509427.5 |
NREP
|
neuronal regeneration related protein |
| chr17_+_45221993 | 0.09 |
ENST00000328118.7
|
FMNL1
|
formin like 1 |
| chr4_+_15002443 | 0.09 |
ENST00000538197.7
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr5_-_111757549 | 0.09 |
ENST00000419114.6
|
NREP
|
neuronal regeneration related protein |
| chr19_+_4343696 | 0.09 |
ENST00000597036.5
|
MPND
|
MPN domain containing |
| chr19_+_1407731 | 0.09 |
ENST00000592453.2
|
DAZAP1
|
DAZ associated protein 1 |
| chr18_+_50967991 | 0.09 |
ENST00000588577.5
|
ELAC1
|
elaC ribonuclease Z 1 |
| chr17_+_37407214 | 0.09 |
ENST00000612272.4
ENST00000621780.1 ENST00000615328.4 |
TADA2A
|
transcriptional adaptor 2A |
| chr17_-_5500997 | 0.09 |
ENST00000568641.2
|
ENSG00000286190.2
|
novel protein |
| chr8_-_92103217 | 0.09 |
ENST00000615601.4
ENST00000523629.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr2_+_30231524 | 0.09 |
ENST00000395323.9
ENST00000406087.5 ENST00000404397.5 |
LBH
|
LBH regulator of WNT signaling pathway |
| chr3_-_133895453 | 0.08 |
ENST00000486858.5
ENST00000477759.5 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr15_+_72118392 | 0.08 |
ENST00000340912.6
|
SENP8
|
SUMO peptidase family member, NEDD8 specific |
| chr12_+_32399517 | 0.08 |
ENST00000534526.7
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr19_+_14433284 | 0.08 |
ENST00000242783.11
|
PKN1
|
protein kinase N1 |
| chr18_-_500692 | 0.08 |
ENST00000400256.5
|
COLEC12
|
collectin subfamily member 12 |
| chr19_+_17933001 | 0.08 |
ENST00000445755.7
|
CCDC124
|
coiled-coil domain containing 124 |
| chr9_-_75088198 | 0.08 |
ENST00000376808.8
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr3_+_49412203 | 0.08 |
ENST00000273590.3
|
TCTA
|
T cell leukemia translocation altered |
| chr4_+_15002541 | 0.08 |
ENST00000507071.6
ENST00000345451.8 ENST00000382395.8 ENST00000382401.8 ENST00000442003.6 |
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr10_+_91798398 | 0.08 |
ENST00000371627.5
|
TNKS2
|
tankyrase 2 |
| chr19_-_3062772 | 0.07 |
ENST00000586742.5
|
TLE5
|
TLE family member 5, transcriptional modulator |
| chr5_-_111757704 | 0.07 |
ENST00000379671.7
|
NREP
|
neuronal regeneration related protein |
| chr19_+_4343527 | 0.07 |
ENST00000262966.12
ENST00000359935.8 ENST00000599840.6 |
MPND
|
MPN domain containing |
| chr3_-_149971109 | 0.07 |
ENST00000239940.11
|
PFN2
|
profilin 2 |
| chr8_-_56446572 | 0.07 |
ENST00000518974.5
ENST00000451791.7 ENST00000523051.5 ENST00000518770.1 |
PENK
|
proenkephalin |
| chr19_+_18419374 | 0.07 |
ENST00000270061.12
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr5_-_111757465 | 0.07 |
ENST00000446294.6
|
NREP
|
neuronal regeneration related protein |
| chr17_+_57256514 | 0.07 |
ENST00000284073.7
ENST00000674964.1 |
MSI2
|
musashi RNA binding protein 2 |
| chr17_+_18183052 | 0.07 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr20_-_63831214 | 0.07 |
ENST00000302995.2
ENST00000245663.9 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr18_-_3013114 | 0.07 |
ENST00000677752.1
|
LPIN2
|
lipin 2 |
| chr22_-_31346317 | 0.07 |
ENST00000266269.10
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr9_+_75088498 | 0.07 |
ENST00000346234.7
|
OSTF1
|
osteoclast stimulating factor 1 |
| chr12_-_31591129 | 0.07 |
ENST00000389082.10
|
DENND5B
|
DENN domain containing 5B |
| chr17_+_57257002 | 0.07 |
ENST00000322684.7
ENST00000579590.5 |
MSI2
|
musashi RNA binding protein 2 |
| chr2_-_55419565 | 0.07 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr11_+_72080595 | 0.07 |
ENST00000647530.1
ENST00000539271.6 ENST00000642510.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr11_-_6419394 | 0.07 |
ENST00000311051.7
|
APBB1
|
amyloid beta precursor protein binding family B member 1 |
| chr5_-_81751085 | 0.07 |
ENST00000515395.5
|
SSBP2
|
single stranded DNA binding protein 2 |
| chr3_-_186109067 | 0.07 |
ENST00000306376.10
|
ETV5
|
ETS variant transcription factor 5 |
| chr9_-_21994345 | 0.07 |
ENST00000579755.2
ENST00000530628.2 |
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr11_+_72080803 | 0.07 |
ENST00000423494.6
ENST00000539587.6 ENST00000536917.2 ENST00000538478.5 ENST00000324866.11 ENST00000643715.1 ENST00000439209.5 |
LRTOMT
ENSG00000284922.2
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chr10_-_79445617 | 0.07 |
ENST00000372336.4
|
ZCCHC24
|
zinc finger CCHC-type containing 24 |
| chrX_-_143635081 | 0.07 |
ENST00000338017.8
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chrX_-_154097668 | 0.07 |
ENST00000407218.5
ENST00000303391.11 ENST00000453960.7 |
MECP2
|
methyl-CpG binding protein 2 |
| chr16_+_1309621 | 0.07 |
ENST00000397515.6
ENST00000397514.8 ENST00000567383.6 ENST00000403747.6 |
UBE2I
|
ubiquitin conjugating enzyme E2 I |
| chr2_-_144516154 | 0.07 |
ENST00000637304.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr9_+_36136703 | 0.07 |
ENST00000377960.9
ENST00000377959.5 |
GLIPR2
|
GLI pathogenesis related 2 |
| chr12_+_94148553 | 0.07 |
ENST00000258526.9
|
PLXNC1
|
plexin C1 |
| chr1_+_65309517 | 0.07 |
ENST00000371069.5
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr20_+_36461303 | 0.07 |
ENST00000475894.5
|
DLGAP4
|
DLG associated protein 4 |
| chr9_+_36136416 | 0.07 |
ENST00000396613.7
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr15_+_91100194 | 0.07 |
ENST00000394232.6
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr9_-_75088140 | 0.07 |
ENST00000361092.9
ENST00000376811.5 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr11_+_65014103 | 0.07 |
ENST00000246747.9
ENST00000529384.5 ENST00000533729.1 |
ARL2
|
ADP ribosylation factor like GTPase 2 |
| chr1_-_161132577 | 0.07 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr3_-_127822455 | 0.07 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr1_+_228487356 | 0.07 |
ENST00000305943.9
|
RNF187
|
ring finger protein 187 |
| chr9_+_36136752 | 0.07 |
ENST00000619700.1
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr2_-_144516397 | 0.07 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_6419051 | 0.06 |
ENST00000299402.10
ENST00000532020.2 ENST00000609360.6 ENST00000389906.6 |
APBB1
|
amyloid beta precursor protein binding family B member 1 |
| chr17_+_21376321 | 0.06 |
ENST00000583088.6
|
KCNJ12
|
potassium inwardly rectifying channel subfamily J member 12 |
| chr19_+_4343587 | 0.06 |
ENST00000596722.5
|
MPND
|
MPN domain containing |
| chr17_-_42276341 | 0.06 |
ENST00000293328.8
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr1_-_145957969 | 0.06 |
ENST00000604000.4
|
LIX1L
|
limb and CNS expressed 1 like |
| chrX_+_136147465 | 0.06 |
ENST00000651929.2
|
FHL1
|
four and a half LIM domains 1 |
| chr18_-_5296139 | 0.06 |
ENST00000400143.7
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr3_-_127823177 | 0.06 |
ENST00000434178.6
|
MGLL
|
monoglyceride lipase |
| chr1_-_28643005 | 0.06 |
ENST00000263974.4
ENST00000373824.9 ENST00000495422.2 |
TAF12
|
TATA-box binding protein associated factor 12 |
| chr10_+_93993897 | 0.06 |
ENST00000371380.8
|
PLCE1
|
phospholipase C epsilon 1 |
| chr17_+_83079595 | 0.06 |
ENST00000320095.12
|
METRNL
|
meteorin like, glial cell differentiation regulator |
| chr18_+_46333956 | 0.06 |
ENST00000587853.1
|
RNF165
|
ring finger protein 165 |
| chr8_+_17497108 | 0.06 |
ENST00000470360.5
|
SLC7A2
|
solute carrier family 7 member 2 |
| chr19_+_3572971 | 0.06 |
ENST00000453933.5
ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr5_-_81751022 | 0.06 |
ENST00000509013.2
ENST00000505980.5 ENST00000509053.5 |
SSBP2
|
single stranded DNA binding protein 2 |
| chrX_+_136497079 | 0.06 |
ENST00000535601.5
ENST00000448450.5 ENST00000425695.5 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr14_+_75428011 | 0.06 |
ENST00000651602.1
ENST00000559060.5 |
JDP2
|
Jun dimerization protein 2 |
| chr19_+_3572777 | 0.06 |
ENST00000416526.5
|
HMG20B
|
high mobility group 20B |
| chr1_+_150149819 | 0.06 |
ENST00000369124.5
|
PLEKHO1
|
pleckstrin homology domain containing O1 |
| chr22_+_30694851 | 0.06 |
ENST00000332585.11
|
OSBP2
|
oxysterol binding protein 2 |
| chr4_-_6709831 | 0.06 |
ENST00000320848.7
|
MRFAP1L1
|
Morf4 family associated protein 1 like 1 |
| chr3_-_127823235 | 0.06 |
ENST00000398104.5
|
MGLL
|
monoglyceride lipase |
| chr4_+_41256921 | 0.06 |
ENST00000284440.9
ENST00000508768.5 ENST00000512788.1 |
UCHL1
|
ubiquitin C-terminal hydrolase L1 |
| chr6_+_117265550 | 0.06 |
ENST00000352536.7
ENST00000326274.6 |
VGLL2
|
vestigial like family member 2 |
| chrX_+_136147556 | 0.06 |
ENST00000651089.1
ENST00000420362.5 |
FHL1
|
four and a half LIM domains 1 |
| chr3_+_143119749 | 0.06 |
ENST00000309575.5
|
CHST2
|
carbohydrate sulfotransferase 2 |
| chr5_+_177446445 | 0.06 |
ENST00000507881.5
|
PRR7
|
proline rich 7, synaptic |
| chr21_+_42974510 | 0.06 |
ENST00000432907.6
ENST00000291547.10 |
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr2_-_55419821 | 0.06 |
ENST00000644630.1
ENST00000471947.2 ENST00000436346.7 ENST00000642200.1 ENST00000413716.7 ENST00000263630.13 ENST00000645072.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr19_+_18419322 | 0.06 |
ENST00000348495.10
|
SSBP4
|
single stranded DNA binding protein 4 |
| chrX_+_136147525 | 0.06 |
ENST00000652745.1
ENST00000627578.2 ENST00000652457.1 ENST00000394155.8 ENST00000618438.4 |
FHL1
|
four and a half LIM domains 1 |
| chr9_+_134326435 | 0.06 |
ENST00000481739.2
|
RXRA
|
retinoid X receptor alpha |
| chr11_+_19712823 | 0.06 |
ENST00000396085.6
ENST00000349880.9 |
NAV2
|
neuron navigator 2 |
| chr11_-_74398378 | 0.06 |
ENST00000298198.5
|
PGM2L1
|
phosphoglucomutase 2 like 1 |
| chr2_+_135741717 | 0.06 |
ENST00000415164.5
|
UBXN4
|
UBX domain protein 4 |
| chr16_+_1309136 | 0.06 |
ENST00000325437.9
|
UBE2I
|
ubiquitin conjugating enzyme E2 I |
| chr5_-_81751103 | 0.06 |
ENST00000514493.5
ENST00000320672.8 ENST00000615665.4 |
SSBP2
|
single stranded DNA binding protein 2 |
| chr8_+_17497078 | 0.06 |
ENST00000494857.6
ENST00000522656.5 |
SLC7A2
|
solute carrier family 7 member 2 |
| chr19_+_18008162 | 0.06 |
ENST00000593560.6
ENST00000222250.5 |
ARRDC2
|
arrestin domain containing 2 |
| chr22_-_38794111 | 0.06 |
ENST00000406622.5
ENST00000216068.9 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein axonemal light chain 4 |
| chr17_-_75182536 | 0.06 |
ENST00000578238.2
|
SUMO2
|
small ubiquitin like modifier 2 |
| chr6_+_71886900 | 0.06 |
ENST00000517960.5
ENST00000518273.5 ENST00000522291.5 ENST00000521978.5 ENST00000520567.5 ENST00000264839.11 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr5_-_169301098 | 0.06 |
ENST00000519560.6
|
SLIT3
|
slit guidance ligand 3 |
| chr1_+_35807974 | 0.05 |
ENST00000373210.4
|
AGO4
|
argonaute RISC component 4 |
| chr10_+_102419189 | 0.05 |
ENST00000432590.5
|
FBXL15
|
F-box and leucine rich repeat protein 15 |
| chr3_-_127822835 | 0.05 |
ENST00000453507.6
|
MGLL
|
monoglyceride lipase |
| chr7_-_74851518 | 0.05 |
ENST00000651129.1
ENST00000614386.1 ENST00000625377.2 ENST00000451013.7 |
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr2_+_30147516 | 0.05 |
ENST00000402708.5
|
YPEL5
|
yippee like 5 |
| chr8_+_84183262 | 0.05 |
ENST00000522455.5
ENST00000521695.5 ENST00000521268.6 |
RALYL
|
RALY RNA binding protein like |
| chr1_-_3531403 | 0.05 |
ENST00000294599.8
|
MEGF6
|
multiple EGF like domains 6 |
| chr11_-_78417788 | 0.05 |
ENST00000361507.5
|
GAB2
|
GRB2 associated binding protein 2 |
| chr14_+_95876762 | 0.05 |
ENST00000503525.2
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA |
| chr7_+_98617275 | 0.05 |
ENST00000265634.4
|
NPTX2
|
neuronal pentraxin 2 |
| chr1_+_42682954 | 0.05 |
ENST00000436427.1
|
YBX1
|
Y-box binding protein 1 |
| chr12_+_123973123 | 0.05 |
ENST00000539644.5
|
ZNF664
|
zinc finger protein 664 |
| chr9_+_101028721 | 0.05 |
ENST00000374874.8
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr4_-_77819356 | 0.05 |
ENST00000649644.1
ENST00000504123.6 ENST00000515441.2 |
CNOT6L
|
CCR4-NOT transcription complex subunit 6 like |
| chr9_+_124257923 | 0.05 |
ENST00000320246.10
ENST00000373600.7 |
NEK6
|
NIMA related kinase 6 |
| chr10_-_931624 | 0.05 |
ENST00000406525.6
|
LARP4B
|
La ribonucleoprotein 4B |
| chr16_+_67279508 | 0.05 |
ENST00000379344.8
ENST00000568621.1 ENST00000450733.5 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chrX_-_154486578 | 0.05 |
ENST00000630530.1
ENST00000369660.9 ENST00000369653.8 |
UBL4A
|
ubiquitin like 4A |
| chr3_-_49411917 | 0.05 |
ENST00000454011.7
ENST00000445425.6 ENST00000422781.6 ENST00000418115.6 ENST00000678921.2 ENST00000676712.2 |
RHOA
|
ras homolog family member A |
| chr14_+_60249191 | 0.05 |
ENST00000395076.9
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A |
| chr3_-_180037019 | 0.05 |
ENST00000485199.5
|
PEX5L
|
peroxisomal biogenesis factor 5 like |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.2 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1903400 | L-arginine transmembrane transport(GO:1903400) |
| 0.0 | 0.1 | GO:0042779 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 0.1 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.0 | GO:0010735 | atrioventricular node development(GO:0003162) positive regulation of transcription via serum response element binding(GO:0010735) His-Purkinje system cell differentiation(GO:0060932) |
| 0.0 | 0.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.0 | 0.0 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.0 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0045588 | isoleucine metabolic process(GO:0006549) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.0 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.1 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.0 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |