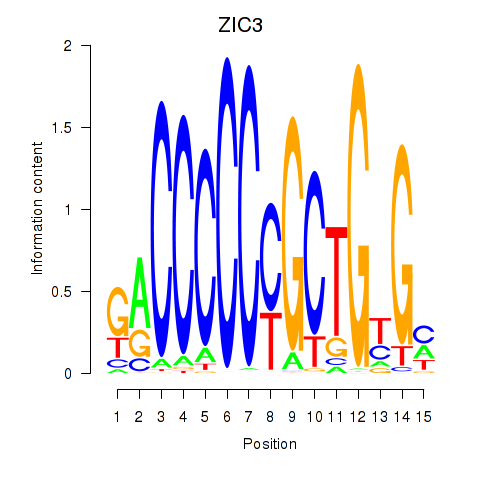
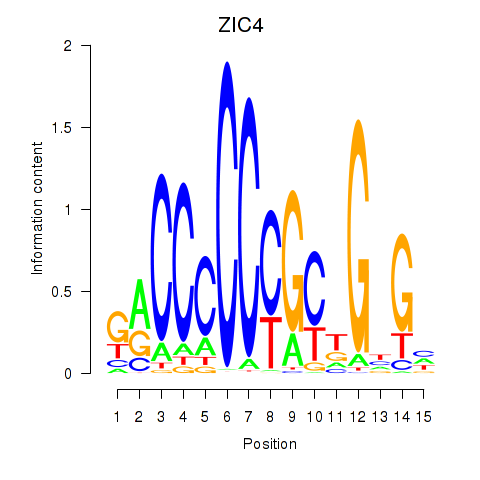
|
chr5_+_76403266
Show fit
|
1.60 |
ENST00000274364.11
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr8_+_1763766
Show fit
|
1.00 |
ENST00000635751.1
ENST00000331222.6
ENST00000637156.1
ENST00000636934.1
ENST00000637083.1
|
CLN8
|
CLN8 transmembrane ER and ERGIC protein
|
|
chr2_-_98936155
Show fit
|
0.98 |
ENST00000428096.5
ENST00000397899.7
ENST00000420294.1
|
CRACDL
|
CRACD like
|
|
chr12_+_55681711
Show fit
|
0.83 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B
|
|
chr12_+_55681647
Show fit
|
0.83 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B
|
|
chr1_+_201648136
Show fit
|
0.75 |
ENST00000367296.8
|
NAV1
|
neuron navigator 1
|
|
chr10_-_13099652
Show fit
|
0.73 |
ENST00000378839.1
|
CCDC3
|
coiled-coil domain containing 3
|
|
chr17_-_44968263
Show fit
|
0.72 |
ENST00000253407.4
|
C1QL1
|
complement C1q like 1
|
|
chr17_-_5500997
Show fit
|
0.67 |
ENST00000568641.2
|
ENSG00000286190.2
|
novel protein
|
|
chr5_+_127290785
Show fit
|
0.62 |
ENST00000503335.7
|
MEGF10
|
multiple EGF like domains 10
|
|
chr7_+_149873956
Show fit
|
0.60 |
ENST00000425642.3
ENST00000479613.5
ENST00000606024.5
ENST00000464662.5
|
ATP6V0E2
|
ATPase H+ transporting V0 subunit e2
|
|
chr4_+_55346349
Show fit
|
0.58 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr2_+_109129199
Show fit
|
0.53 |
ENST00000309415.8
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr8_+_1763752
Show fit
|
0.53 |
ENST00000519254.2
|
CLN8
|
CLN8 transmembrane ER and ERGIC protein
|
|
chr16_+_30985181
Show fit
|
0.52 |
ENST00000262520.10
ENST00000297679.10
ENST00000562932.5
ENST00000574447.1
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr8_+_1763832
Show fit
|
0.51 |
ENST00000520991.3
|
CLN8
|
CLN8 transmembrane ER and ERGIC protein
|
|
chr17_+_80220406
Show fit
|
0.51 |
ENST00000573809.5
ENST00000361193.8
ENST00000574967.5
ENST00000576126.5
ENST00000411502.7
ENST00000546047.6
ENST00000572725.5
|
SLC26A11
|
solute carrier family 26 member 11
|
|
chr4_+_15002443
Show fit
|
0.49 |
ENST00000538197.7
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr4_+_15002541
Show fit
|
0.48 |
ENST00000507071.6
ENST00000345451.8
ENST00000382395.8
ENST00000382401.8
ENST00000442003.6
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr2_+_23385170
Show fit
|
0.48 |
ENST00000486442.6
|
KLHL29
|
kelch like family member 29
|
|
chr3_-_112638097
Show fit
|
0.47 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr5_+_127290806
Show fit
|
0.47 |
ENST00000508365.5
ENST00000418761.6
ENST00000274473.6
|
MEGF10
|
multiple EGF like domains 10
|
|
chr21_+_36135071
Show fit
|
0.47 |
ENST00000290354.6
|
CBR3
|
carbonyl reductase 3
|
|
chr10_+_13100075
Show fit
|
0.45 |
ENST00000378747.8
ENST00000378757.6
ENST00000378752.7
ENST00000378748.7
|
OPTN
|
optineurin
|
|
chr1_+_89524871
Show fit
|
0.44 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B
|
|
chr1_+_89524819
Show fit
|
0.44 |
ENST00000439853.6
ENST00000330947.7
ENST00000449440.5
ENST00000640258.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B
|
|
chr17_-_80220325
Show fit
|
0.44 |
ENST00000326317.11
ENST00000570427.1
ENST00000570923.1
|
SGSH
|
N-sulfoglucosamine sulfohydrolase
|
|
chr18_-_28177934
Show fit
|
0.44 |
ENST00000676445.1
|
CDH2
|
cadherin 2
|
|
chr10_-_77637558
Show fit
|
0.42 |
ENST00000372421.10
ENST00000639370.1
ENST00000640773.1
ENST00000638895.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr3_+_12289061
Show fit
|
0.42 |
ENST00000652522.1
ENST00000652431.1
ENST00000652098.1
ENST00000651735.1
ENST00000397026.7
|
PPARG
|
peroxisome proliferator activated receptor gamma
|
|
chr1_+_215082731
Show fit
|
0.41 |
ENST00000444842.7
|
KCNK2
|
potassium two pore domain channel subfamily K member 2
|
|
chr3_+_12288838
Show fit
|
0.41 |
ENST00000455517.6
ENST00000681982.1
|
PPARG
|
peroxisome proliferator activated receptor gamma
|
|
chr19_-_31349408
Show fit
|
0.41 |
ENST00000240587.5
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr5_-_9546066
Show fit
|
0.40 |
ENST00000382496.10
ENST00000652226.1
|
SEMA5A
|
semaphorin 5A
|
|
chr10_-_77638369
Show fit
|
0.39 |
ENST00000372443.6
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr12_+_49961864
Show fit
|
0.37 |
ENST00000293599.7
|
AQP5
|
aquaporin 5
|
|
chr10_-_77637902
Show fit
|
0.37 |
ENST00000286627.10
ENST00000639486.1
ENST00000640523.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr19_-_58098203
Show fit
|
0.37 |
ENST00000600845.1
ENST00000240727.10
ENST00000600897.5
ENST00000421612.6
ENST00000601063.1
ENST00000601144.6
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr2_-_45009401
Show fit
|
0.35 |
ENST00000303077.7
|
SIX2
|
SIX homeobox 2
|
|
chr10_-_77637444
Show fit
|
0.35 |
ENST00000639205.1
ENST00000639498.1
ENST00000372408.7
ENST00000372403.9
ENST00000640626.1
ENST00000404857.6
ENST00000638252.1
ENST00000640029.1
ENST00000640934.1
ENST00000639823.1
ENST00000372437.6
ENST00000639344.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr13_+_34942263
Show fit
|
0.34 |
ENST00000379939.7
ENST00000400445.7
|
NBEA
|
neurobeachin
|
|
chr1_-_212699817
Show fit
|
0.34 |
ENST00000243440.2
|
BATF3
|
basic leucine zipper ATF-like transcription factor 3
|
|
chr2_+_127423265
Show fit
|
0.34 |
ENST00000402125.2
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa
|
|
chr6_-_88166339
Show fit
|
0.33 |
ENST00000369501.3
ENST00000551417.2
|
CNR1
|
cannabinoid receptor 1
|
|
chr6_+_33200860
Show fit
|
0.31 |
ENST00000374677.8
|
SLC39A7
|
solute carrier family 39 member 7
|
|
chr10_-_77637789
Show fit
|
0.31 |
ENST00000481070.1
ENST00000640969.1
ENST00000286628.14
ENST00000638991.1
ENST00000639913.1
ENST00000480683.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr20_+_3888915
Show fit
|
0.31 |
ENST00000316562.9
ENST00000495692.5
|
PANK2
|
pantothenate kinase 2
|
|
chr6_+_33200820
Show fit
|
0.31 |
ENST00000374675.7
|
SLC39A7
|
solute carrier family 39 member 7
|
|
chr10_-_77637721
Show fit
|
0.30 |
ENST00000638848.1
ENST00000639406.1
ENST00000618048.2
ENST00000639120.1
ENST00000640834.1
ENST00000639601.1
ENST00000638514.1
ENST00000457953.6
ENST00000639090.1
ENST00000639489.1
ENST00000372440.6
ENST00000404771.8
ENST00000638203.1
ENST00000638306.1
ENST00000638351.1
ENST00000638606.1
ENST00000639591.1
ENST00000640182.1
ENST00000640605.1
ENST00000640141.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr10_+_13099585
Show fit
|
0.30 |
ENST00000378764.6
|
OPTN
|
optineurin
|
|
chr14_+_100381974
Show fit
|
0.30 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25
|
|
chr17_+_40443441
Show fit
|
0.29 |
ENST00000269593.5
|
IGFBP4
|
insulin like growth factor binding protein 4
|
|
chr20_-_57711536
Show fit
|
0.28 |
ENST00000265626.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr19_+_36605292
Show fit
|
0.27 |
ENST00000460670.5
ENST00000292928.7
ENST00000439428.5
|
ZNF382
|
zinc finger protein 382
|
|
chr5_-_111756245
Show fit
|
0.27 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein
|
|
chr2_+_20447065
Show fit
|
0.27 |
ENST00000272233.6
|
RHOB
|
ras homolog family member B
|
|
chr5_-_14871757
Show fit
|
0.25 |
ENST00000284268.8
|
ANKH
|
ANKH inorganic pyrophosphate transport regulator
|
|
chrX_+_153517672
Show fit
|
0.25 |
ENST00000349466.6
ENST00000370186.5
ENST00000359149.8
|
ATP2B3
|
ATPase plasma membrane Ca2+ transporting 3
|
|
chr12_+_51391273
Show fit
|
0.25 |
ENST00000535225.6
ENST00000358657.7
|
SLC4A8
|
solute carrier family 4 member 8
|
|
chr10_+_84194621
Show fit
|
0.25 |
ENST00000332904.7
|
CDHR1
|
cadherin related family member 1
|
|
chr10_+_13099440
Show fit
|
0.25 |
ENST00000263036.9
|
OPTN
|
optineurin
|
|
chr20_-_41366805
Show fit
|
0.24 |
ENST00000332312.4
|
EMILIN3
|
elastin microfibril interfacer 3
|
|
chrX_+_154458274
Show fit
|
0.24 |
ENST00000369682.4
|
PLXNA3
|
plexin A3
|
|
chr11_-_27700447
Show fit
|
0.24 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor
|
|
chr5_+_141373878
Show fit
|
0.24 |
ENST00000517434.3
ENST00000610583.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr19_+_49114324
Show fit
|
0.23 |
ENST00000391864.7
|
LIN7B
|
lin-7 homolog B, crumbs cell polarity complex component
|
|
chr11_-_74398378
Show fit
|
0.23 |
ENST00000298198.5
|
PGM2L1
|
phosphoglucomutase 2 like 1
|
|
chr1_-_42766978
Show fit
|
0.23 |
ENST00000372526.2
ENST00000236040.8
ENST00000296388.10
ENST00000397054.7
|
P3H1
|
prolyl 3-hydroxylase 1
|
|
chr16_-_49856105
Show fit
|
0.23 |
ENST00000563137.7
|
ZNF423
|
zinc finger protein 423
|
|
chr17_+_76737387
Show fit
|
0.23 |
ENST00000590393.1
ENST00000355954.7
ENST00000586689.5
ENST00000587661.5
ENST00000593181.5
ENST00000336509.8
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr6_-_38639898
Show fit
|
0.22 |
ENST00000481247.6
|
BTBD9
|
BTB domain containing 9
|
|
chr17_-_41034871
Show fit
|
0.22 |
ENST00000344363.7
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr12_-_48004496
Show fit
|
0.22 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain
|
|
chr7_-_50782853
Show fit
|
0.22 |
ENST00000401949.6
|
GRB10
|
growth factor receptor bound protein 10
|
|
chr5_+_176810498
Show fit
|
0.22 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A
|
|
chr19_+_36604817
Show fit
|
0.22 |
ENST00000423582.5
|
ZNF382
|
zinc finger protein 382
|
|
chr11_-_67505323
Show fit
|
0.22 |
ENST00000356404.8
ENST00000436757.6
|
PITPNM1
|
phosphatidylinositol transfer protein membrane associated 1
|
|
chrX_-_34657274
Show fit
|
0.21 |
ENST00000275954.4
|
TMEM47
|
transmembrane protein 47
|
|
chr6_-_84763437
Show fit
|
0.21 |
ENST00000606784.5
ENST00000606325.5
|
TBX18
|
T-box transcription factor 18
|
|
chr7_-_117873420
Show fit
|
0.21 |
ENST00000160373.8
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr5_+_176810552
Show fit
|
0.21 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A
|
|
chr1_-_30908681
Show fit
|
0.20 |
ENST00000339394.7
|
SDC3
|
syndecan 3
|
|
chr17_+_40062810
Show fit
|
0.20 |
ENST00000584985.5
ENST00000264637.8
|
THRA
|
thyroid hormone receptor alpha
|
|
chr8_-_27605271
Show fit
|
0.19 |
ENST00000522098.1
|
CLU
|
clusterin
|
|
chr11_-_2161158
Show fit
|
0.19 |
ENST00000421783.1
ENST00000397262.5
ENST00000381330.5
ENST00000250971.7
ENST00000397270.1
|
INS
INS-IGF2
|
insulin
INS-IGF2 readthrough
|
|
chr3_-_46882106
Show fit
|
0.19 |
ENST00000662933.1
|
MYL3
|
myosin light chain 3
|
|
chr2_+_108534353
Show fit
|
0.19 |
ENST00000544547.5
|
LIMS1
|
LIM zinc finger domain containing 1
|
|
chr4_+_55346213
Show fit
|
0.19 |
ENST00000679836.1
ENST00000264228.9
ENST00000679707.1
|
SRD5A3
ENSG00000288695.1
|
steroid 5 alpha-reductase 3
novel protein, SRD5A3-RP11-177J6.1 readthrough
|
|
chr7_-_23347704
Show fit
|
0.19 |
ENST00000619562.4
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3
|
|
chr8_+_22165140
Show fit
|
0.19 |
ENST00000397814.7
ENST00000354870.5
|
BMP1
|
bone morphogenetic protein 1
|
|
chr20_+_43514426
Show fit
|
0.19 |
ENST00000422861.3
|
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1
|
|
chr16_-_18376839
Show fit
|
0.18 |
ENST00000427999.6
|
NPIPA9
|
nuclear pore complex interacting protein family, member A9
|
|
chr6_-_41705813
Show fit
|
0.18 |
ENST00000419574.6
ENST00000445214.2
|
TFEB
|
transcription factor EB
|
|
chr5_-_38556625
Show fit
|
0.18 |
ENST00000506990.5
ENST00000453190.7
|
LIFR
|
LIF receptor subunit alpha
|
|
chr12_-_58920465
Show fit
|
0.18 |
ENST00000320743.8
|
LRIG3
|
leucine rich repeats and immunoglobulin like domains 3
|
|
chr17_-_41027198
Show fit
|
0.18 |
ENST00000361883.6
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr9_+_128420812
Show fit
|
0.18 |
ENST00000372838.9
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr13_-_98752619
Show fit
|
0.17 |
ENST00000376503.10
|
SLC15A1
|
solute carrier family 15 member 1
|
|
chr11_-_6320494
Show fit
|
0.17 |
ENST00000303927.4
ENST00000530979.1
|
CAVIN3
|
caveolae associated protein 3
|
|
chr19_+_49114352
Show fit
|
0.17 |
ENST00000221459.7
ENST00000486217.2
|
LIN7B
|
lin-7 homolog B, crumbs cell polarity complex component
|
|
chr2_+_64989343
Show fit
|
0.17 |
ENST00000234256.4
|
SLC1A4
|
solute carrier family 1 member 4
|
|
chr11_-_12008584
Show fit
|
0.17 |
ENST00000534511.5
ENST00000683431.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3
|
|
chr5_-_157575767
Show fit
|
0.16 |
ENST00000257527.9
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr14_+_105314711
Show fit
|
0.16 |
ENST00000447393.6
ENST00000547217.5
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr14_+_24161257
Show fit
|
0.16 |
ENST00000396864.8
ENST00000557894.5
ENST00000559284.5
ENST00000560275.5
|
IRF9
|
interferon regulatory factor 9
|
|
chr11_-_65382632
Show fit
|
0.16 |
ENST00000294187.10
ENST00000398802.6
ENST00000530936.1
|
SLC25A45
|
solute carrier family 25 member 45
|
|
chr20_+_35615812
Show fit
|
0.16 |
ENST00000679710.1
ENST00000374273.8
|
SPAG4
|
sperm associated antigen 4
|
|
chr7_-_50450324
Show fit
|
0.16 |
ENST00000356889.8
ENST00000420829.5
ENST00000448788.1
ENST00000395556.6
ENST00000433017.6
ENST00000422854.5
ENST00000435566.5
ENST00000617389.4
ENST00000611938.4
ENST00000615084.4
|
FIGNL1
|
fidgetin like 1
|
|
chr19_+_1266653
Show fit
|
0.16 |
ENST00000586472.5
ENST00000589266.5
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr5_+_141338753
Show fit
|
0.16 |
ENST00000528330.2
ENST00000394576.3
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr2_+_24491239
Show fit
|
0.16 |
ENST00000348332.8
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr12_-_98644733
Show fit
|
0.16 |
ENST00000299157.5
ENST00000393042.3
|
IKBIP
|
IKBKB interacting protein
|
|
chr13_-_23433676
Show fit
|
0.16 |
ENST00000682547.1
ENST00000455470.6
ENST00000382292.9
|
SACS
|
sacsin molecular chaperone
|
|
chr17_+_20009320
Show fit
|
0.15 |
ENST00000679049.1
ENST00000472876.5
ENST00000583528.6
ENST00000680019.1
ENST00000681593.1
ENST00000395527.8
ENST00000583482.7
ENST00000679255.1
ENST00000681202.1
ENST00000677914.1
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr20_-_49278034
Show fit
|
0.15 |
ENST00000371744.5
ENST00000396105.6
ENST00000371752.5
|
ZNFX1
|
zinc finger NFX1-type containing 1
|
|
chr7_-_134316912
Show fit
|
0.15 |
ENST00000378509.9
|
SLC35B4
|
solute carrier family 35 member B4
|
|
chr1_+_218346235
Show fit
|
0.15 |
ENST00000366929.4
|
TGFB2
|
transforming growth factor beta 2
|
|
chr10_-_87094761
Show fit
|
0.15 |
ENST00000684338.1
ENST00000684201.1
ENST00000277865.5
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr11_-_67373584
Show fit
|
0.15 |
ENST00000543494.1
ENST00000312438.8
|
ENSG00000256514.1
CLCF1
|
novel protein
cardiotrophin like cytokine factor 1
|
|
chr18_+_50560070
Show fit
|
0.15 |
ENST00000400384.7
ENST00000540640.3
ENST00000592595.5
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr4_+_73853290
Show fit
|
0.15 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1
|
|
chr1_+_52056284
Show fit
|
0.15 |
ENST00000313334.13
ENST00000472944.6
ENST00000484036.1
|
BTF3L4
|
basic transcription factor 3 like 4
|
|
chr4_+_38867789
Show fit
|
0.15 |
ENST00000358869.5
|
FAM114A1
|
family with sequence similarity 114 member A1
|
|
chr4_-_128288791
Show fit
|
0.15 |
ENST00000613358.4
ENST00000520121.6
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr1_-_37034492
Show fit
|
0.15 |
ENST00000373091.8
|
GRIK3
|
glutamate ionotropic receptor kainate type subunit 3
|
|
chr6_-_38639852
Show fit
|
0.14 |
ENST00000498633.1
ENST00000649492.1
|
BTBD9
|
BTB domain containing 9
|
|
chr22_-_41446777
Show fit
|
0.14 |
ENST00000434408.1
ENST00000327492.4
|
TOB2
|
transducer of ERBB2, 2
|
|
chr4_+_38867677
Show fit
|
0.14 |
ENST00000510213.5
ENST00000515037.5
|
FAM114A1
|
family with sequence similarity 114 member A1
|
|
chr19_-_9913819
Show fit
|
0.14 |
ENST00000593091.2
|
OLFM2
|
olfactomedin 2
|
|
chr9_-_111484353
Show fit
|
0.14 |
ENST00000338205.9
ENST00000684092.1
|
ECPAS
|
Ecm29 proteasome adaptor and scaffold
|
|
chr4_-_128287785
Show fit
|
0.14 |
ENST00000296425.10
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr19_+_1908258
Show fit
|
0.14 |
ENST00000411971.5
ENST00000588907.2
|
SCAMP4
|
secretory carrier membrane protein 4
|
|
chr16_-_800705
Show fit
|
0.14 |
ENST00000248150.5
|
GNG13
|
G protein subunit gamma 13
|
|
chr17_+_79034185
Show fit
|
0.14 |
ENST00000581774.5
|
C1QTNF1
|
C1q and TNF related 1
|
|
chr20_+_3888772
Show fit
|
0.14 |
ENST00000497424.5
|
PANK2
|
pantothenate kinase 2
|
|
chr8_+_43140405
Show fit
|
0.14 |
ENST00000379644.9
|
HGSNAT
|
heparan-alpha-glucosaminide N-acetyltransferase
|
|
chr19_+_17967072
Show fit
|
0.14 |
ENST00000684775.1
ENST00000615435.2
ENST00000683588.1
ENST00000684725.1
ENST00000682421.1
|
KCNN1
|
potassium calcium-activated channel subfamily N member 1
|
|
chr2_+_37344594
Show fit
|
0.14 |
ENST00000404976.5
ENST00000338415.8
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr11_+_65014103
Show fit
|
0.14 |
ENST00000246747.9
ENST00000529384.5
ENST00000533729.1
|
ARL2
|
ADP ribosylation factor like GTPase 2
|
|
chr16_+_28985043
Show fit
|
0.13 |
ENST00000395456.7
ENST00000564277.5
ENST00000630764.2
ENST00000354453.7
|
LAT
|
linker for activation of T cells
|
|
chr21_+_44697172
Show fit
|
0.13 |
ENST00000400365.3
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr4_+_2963580
Show fit
|
0.13 |
ENST00000398051.8
ENST00000503518.2
|
GRK4
|
G protein-coupled receptor kinase 4
|
|
chr10_+_28677487
Show fit
|
0.13 |
ENST00000375533.6
|
BAMBI
|
BMP and activin membrane bound inhibitor
|
|
chrX_+_24693879
Show fit
|
0.13 |
ENST00000379068.8
ENST00000677890.1
ENST00000379059.7
|
POLA1
|
DNA polymerase alpha 1, catalytic subunit
|
|
chr9_+_136980211
Show fit
|
0.13 |
ENST00000444903.2
|
PTGDS
|
prostaglandin D2 synthase
|
|
chr14_-_23352741
Show fit
|
0.13 |
ENST00000354772.9
|
SLC22A17
|
solute carrier family 22 member 17
|
|
chr8_+_17156463
Show fit
|
0.13 |
ENST00000262096.13
|
ZDHHC2
|
zinc finger DHHC-type palmitoyltransferase 2
|
|
chr9_-_83063159
Show fit
|
0.13 |
ENST00000340717.4
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr2_-_97995916
Show fit
|
0.13 |
ENST00000186436.10
|
TMEM131
|
transmembrane protein 131
|
|
chr22_-_41240925
Show fit
|
0.13 |
ENST00000216241.14
|
CHADL
|
chondroadherin like
|
|
chr3_-_64268161
Show fit
|
0.13 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2
|
|
chr19_-_2151525
Show fit
|
0.13 |
ENST00000345016.9
ENST00000643116.3
|
AP3D1
|
adaptor related protein complex 3 subunit delta 1
|
|
chr19_+_33194308
Show fit
|
0.13 |
ENST00000253193.9
|
LRP3
|
LDL receptor related protein 3
|
|
chr5_+_134371561
Show fit
|
0.13 |
ENST00000265339.7
ENST00000506787.5
ENST00000507277.1
|
UBE2B
|
ubiquitin conjugating enzyme E2 B
|
|
chr20_+_58840692
Show fit
|
0.13 |
ENST00000306090.12
|
GNAS
|
GNAS complex locus
|
|
chr9_+_19230435
Show fit
|
0.13 |
ENST00000602925.5
|
DENND4C
|
DENN domain containing 4C
|
|
chr19_-_48249841
Show fit
|
0.13 |
ENST00000447740.6
|
CARD8
|
caspase recruitment domain family member 8
|
|
chr4_+_52051285
Show fit
|
0.12 |
ENST00000295213.9
ENST00000419395.6
|
SPATA18
|
spermatogenesis associated 18
|
|
chr3_-_9769910
Show fit
|
0.12 |
ENST00000256460.8
|
CAMK1
|
calcium/calmodulin dependent protein kinase I
|
|
chr2_-_200510044
Show fit
|
0.12 |
ENST00000359878.8
ENST00000409157.5
|
KCTD18
|
potassium channel tetramerization domain containing 18
|
|
chr4_-_21948733
Show fit
|
0.12 |
ENST00000447367.6
ENST00000382152.7
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4
|
|
chr16_+_28985251
Show fit
|
0.12 |
ENST00000360872.9
ENST00000566177.5
|
LAT
|
linker for activation of T cells
|
|
chrX_+_153517626
Show fit
|
0.12 |
ENST00000263519.5
|
ATP2B3
|
ATPase plasma membrane Ca2+ transporting 3
|
|
chr10_-_98268186
Show fit
|
0.12 |
ENST00000260702.4
|
LOXL4
|
lysyl oxidase like 4
|
|
chr9_+_137077467
Show fit
|
0.12 |
ENST00000409858.8
|
UAP1L1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 like 1
|
|
chr17_-_39688016
Show fit
|
0.12 |
ENST00000579146.5
ENST00000300658.9
ENST00000378011.8
ENST00000429199.6
|
PGAP3
|
post-GPI attachment to proteins phospholipase 3
|
|
chr20_-_33686371
Show fit
|
0.12 |
ENST00000343380.6
|
E2F1
|
E2F transcription factor 1
|
|
chr11_-_3165264
Show fit
|
0.12 |
ENST00000389989.7
ENST00000263650.12
|
OSBPL5
|
oxysterol binding protein like 5
|
|
chr3_-_177197429
Show fit
|
0.11 |
ENST00000457928.7
|
TBL1XR1
|
TBL1X receptor 1
|
|
chr2_+_100820102
Show fit
|
0.11 |
ENST00000335681.10
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr1_-_182672232
Show fit
|
0.11 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8
|
|
chr5_-_157575741
Show fit
|
0.11 |
ENST00000517905.1
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr17_+_28728781
Show fit
|
0.11 |
ENST00000268766.11
|
NEK8
|
NIMA related kinase 8
|
|
chr14_-_23352872
Show fit
|
0.11 |
ENST00000397267.5
|
SLC22A17
|
solute carrier family 22 member 17
|
|
chr4_+_114598738
Show fit
|
0.11 |
ENST00000507710.1
ENST00000310836.11
|
UGT8
|
UDP glycosyltransferase 8
|
|
chr7_-_150323489
Show fit
|
0.11 |
ENST00000683684.1
ENST00000478393.5
|
ACTR3C
|
actin related protein 3C
|
|
chrX_-_129843388
Show fit
|
0.11 |
ENST00000371064.7
|
ZDHHC9
|
zinc finger DHHC-type palmitoyltransferase 9
|
|
chr17_-_10729973
Show fit
|
0.11 |
ENST00000578345.1
ENST00000341871.8
ENST00000455996.6
|
TMEM220
|
transmembrane protein 220
|
|
chr9_+_97412062
Show fit
|
0.11 |
ENST00000355295.5
|
TDRD7
|
tudor domain containing 7
|
|
chr7_+_37848953
Show fit
|
0.11 |
ENST00000444718.5
ENST00000440017.5
|
NME8
|
NME/NM23 family member 8
|
|
chr4_-_170089930
Show fit
|
0.11 |
ENST00000337664.9
|
AADAT
|
aminoadipate aminotransferase
|
|
chr21_-_43776238
Show fit
|
0.10 |
ENST00000639959.1
ENST00000291568.7
ENST00000640406.1
|
CSTB
|
cystatin B
|
|
chr3_-_149086488
Show fit
|
0.10 |
ENST00000392912.6
ENST00000465259.5
ENST00000310053.10
ENST00000494055.5
|
HLTF
|
helicase like transcription factor
|
|
chr22_-_46738205
Show fit
|
0.10 |
ENST00000216264.13
|
CERK
|
ceramide kinase
|
|
chr1_-_223845894
Show fit
|
0.10 |
ENST00000391878.6
ENST00000343537.12
|
TP53BP2
|
tumor protein p53 binding protein 2
|
|
chr20_-_63627049
Show fit
|
0.10 |
ENST00000370077.2
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr8_-_27992624
Show fit
|
0.10 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5
|
|
chr1_+_32886456
Show fit
|
0.10 |
ENST00000373467.4
|
HPCA
|
hippocalcin
|
|
chr11_+_1223053
Show fit
|
0.10 |
ENST00000529681.5
|
MUC5B
|
mucin 5B, oligomeric mucus/gel-forming
|
|
chr6_+_144583198
Show fit
|
0.10 |
ENST00000367526.8
|
UTRN
|
utrophin
|
|
chr17_-_19716604
Show fit
|
0.10 |
ENST00000325411.9
ENST00000350657.9
|
SLC47A2
|
solute carrier family 47 member 2
|
|
chr5_-_177497561
Show fit
|
0.10 |
ENST00000359895.6
ENST00000355572.6
ENST00000393551.5
ENST00000505074.5
ENST00000393546.8
ENST00000355841.7
|
PDLIM7
|
PDZ and LIM domain 7
|
|
chr17_-_51120855
Show fit
|
0.10 |
ENST00000618113.4
ENST00000357122.8
ENST00000262013.12
|
SPAG9
|
sperm associated antigen 9
|
|
chr8_-_140457719
Show fit
|
0.10 |
ENST00000438773.4
|
TRAPPC9
|
trafficking protein particle complex 9
|
|
chr17_-_76141240
Show fit
|
0.10 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1
|
|
chr7_+_37848597
Show fit
|
0.10 |
ENST00000199447.9
ENST00000455500.5
|
NME8
|
NME/NM23 family member 8
|
|
chr14_-_102509798
Show fit
|
0.10 |
ENST00000560748.5
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr11_+_73308237
Show fit
|
0.10 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17
|
|
chr3_-_86991135
Show fit
|
0.09 |
ENST00000398399.7
|
VGLL3
|
vestigial like family member 3
|
|
chr8_+_67064316
Show fit
|
0.09 |
ENST00000675306.2
ENST00000678017.1
ENST00000262210.11
ENST00000675869.1
ENST00000677009.1
ENST00000676847.1
ENST00000676471.1
ENST00000678542.1
ENST00000677619.1
ENST00000676605.1
ENST00000678553.1
ENST00000674993.1
ENST00000678318.1
ENST00000676573.1
ENST00000676317.1
ENST00000677592.1
ENST00000679226.1
ENST00000675955.1
ENST00000676882.1
ENST00000678616.1
ENST00000678645.1
ENST00000678747.1
|
CSPP1
|
centrosome and spindle pole associated protein 1
|