Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
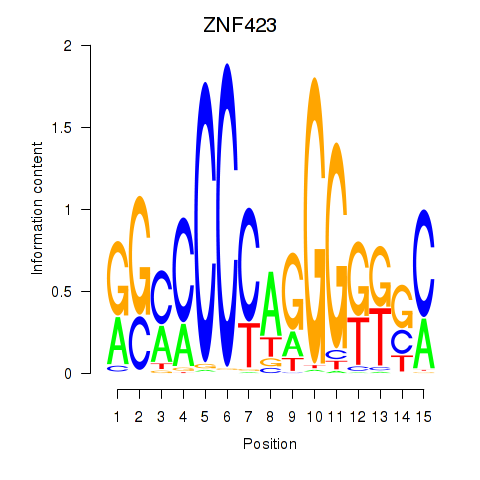
Results for ZNF423
Z-value: 0.73
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.12 | zinc finger protein 423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF423 | hg38_v1_chr16_-_49664225_49664298, hg38_v1_chr16_-_49856105_49856119 | -0.10 | 8.1e-01 | Click! |
Activity profile of ZNF423 motif
Sorted Z-values of ZNF423 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_53480619 | 1.02 |
ENST00000396947.7
ENST00000256759.8 |
FST
|
follistatin |
| chr7_-_44225893 | 0.93 |
ENST00000425809.5
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta |
| chr6_+_30884063 | 0.57 |
ENST00000511510.5
ENST00000376569.7 ENST00000376570.8 ENST00000504927.5 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_54385727 | 0.47 |
ENST00000551109.5
ENST00000546970.5 |
ZNF385A
|
zinc finger protein 385A |
| chr11_+_60924452 | 0.46 |
ENST00000453848.7
ENST00000544065.5 ENST00000005286.8 |
TMEM132A
|
transmembrane protein 132A |
| chr16_+_2988256 | 0.43 |
ENST00000573315.2
|
LINC00514
|
long intergenic non-protein coding RNA 514 |
| chr5_+_69492767 | 0.41 |
ENST00000681041.1
ENST00000680098.1 ENST00000680784.1 ENST00000396442.7 ENST00000681895.1 |
OCLN
|
occludin |
| chr9_+_113876282 | 0.41 |
ENST00000374126.9
ENST00000615615.4 ENST00000288466.11 |
ZNF618
|
zinc finger protein 618 |
| chr7_+_20330893 | 0.40 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr7_+_20330678 | 0.36 |
ENST00000537992.5
|
ITGB8
|
integrin subunit beta 8 |
| chr11_-_44950867 | 0.36 |
ENST00000528290.5
ENST00000525680.6 ENST00000530035.5 ENST00000527685.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr1_-_32901330 | 0.36 |
ENST00000329151.5
ENST00000373463.8 |
TMEM54
|
transmembrane protein 54 |
| chr11_-_44950839 | 0.33 |
ENST00000395648.7
ENST00000531928.6 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr8_-_79767843 | 0.33 |
ENST00000337919.9
|
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1 |
| chr11_+_45922640 | 0.32 |
ENST00000401752.6
ENST00000325468.9 |
LARGE2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr19_-_11578937 | 0.32 |
ENST00000592659.1
ENST00000592828.6 ENST00000218758.9 ENST00000412435.6 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr8_-_79767462 | 0.30 |
ENST00000674295.1
ENST00000518733.1 ENST00000674418.1 ENST00000674358.1 ENST00000354724.8 |
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1 |
| chr16_+_2964216 | 0.30 |
ENST00000572045.5
ENST00000571007.5 ENST00000575885.5 ENST00000303746.10 ENST00000319500.10 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr16_+_3046552 | 0.29 |
ENST00000336577.9
|
MMP25
|
matrix metallopeptidase 25 |
| chr7_-_19145306 | 0.29 |
ENST00000275461.3
|
FERD3L
|
Fer3 like bHLH transcription factor |
| chr11_-_44950151 | 0.28 |
ENST00000533940.5
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr9_+_109780179 | 0.27 |
ENST00000314527.9
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr17_-_29180359 | 0.26 |
ENST00000531253.5
ENST00000628822.2 |
MYO18A
|
myosin XVIIIA |
| chrX_-_48971829 | 0.26 |
ENST00000218176.4
|
KCND1
|
potassium voltage-gated channel subfamily D member 1 |
| chr17_-_29180377 | 0.26 |
ENST00000527372.7
|
MYO18A
|
myosin XVIIIA |
| chr9_+_98807619 | 0.26 |
ENST00000375011.4
|
GALNT12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr8_-_42377227 | 0.25 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chrX_-_51496572 | 0.24 |
ENST00000375992.4
|
NUDT11
|
nudix hydrolase 11 |
| chr16_+_27313879 | 0.23 |
ENST00000562142.5
ENST00000561742.5 ENST00000543915.6 ENST00000395762.7 ENST00000563002.5 |
IL4R
|
interleukin 4 receptor |
| chr9_-_39239174 | 0.22 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr16_-_11587450 | 0.20 |
ENST00000571688.5
|
LITAF
|
lipopolysaccharide induced TNF factor |
| chr5_+_171309239 | 0.19 |
ENST00000296921.6
|
TLX3
|
T cell leukemia homeobox 3 |
| chr5_+_136049513 | 0.19 |
ENST00000514554.5
|
TGFBI
|
transforming growth factor beta induced |
| chr12_+_109713817 | 0.18 |
ENST00000538780.2
|
FAM222A
|
family with sequence similarity 222 member A |
| chr16_-_11587162 | 0.18 |
ENST00000570904.5
ENST00000574701.5 |
LITAF
|
lipopolysaccharide induced TNF factor |
| chr19_+_45251249 | 0.18 |
ENST00000262891.9
ENST00000300843.8 |
MARK4
|
microtubule affinity regulating kinase 4 |
| chr2_+_27496830 | 0.18 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator |
| chr6_+_33621313 | 0.18 |
ENST00000605930.3
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor type 3 |
| chr19_+_45251363 | 0.18 |
ENST00000620044.4
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr3_-_196082078 | 0.17 |
ENST00000360110.9
ENST00000392396.7 ENST00000420415.5 |
TFRC
|
transferrin receptor |
| chr16_-_70685975 | 0.17 |
ENST00000338779.11
|
MTSS2
|
MTSS I-BAR domain containing 2 |
| chr5_-_176630364 | 0.16 |
ENST00000310112.7
|
SNCB
|
synuclein beta |
| chr10_+_84173793 | 0.16 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr6_+_109440695 | 0.16 |
ENST00000258052.8
|
SMPD2
|
sphingomyelin phosphodiesterase 2 |
| chr3_+_6861107 | 0.15 |
ENST00000357716.9
ENST00000486284.5 ENST00000389336.8 |
GRM7
|
glutamate metabotropic receptor 7 |
| chr12_+_7189582 | 0.15 |
ENST00000266563.9
ENST00000543974.5 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr10_+_133527355 | 0.15 |
ENST00000252945.8
ENST00000421586.5 ENST00000418356.1 |
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1 |
| chr10_+_125896549 | 0.15 |
ENST00000368693.6
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr5_-_176630517 | 0.15 |
ENST00000393693.7
ENST00000614675.4 |
SNCB
|
synuclein beta |
| chr12_+_7189845 | 0.14 |
ENST00000412720.6
ENST00000396637.7 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr4_-_119627631 | 0.14 |
ENST00000264805.9
|
PDE5A
|
phosphodiesterase 5A |
| chr6_-_142147122 | 0.14 |
ENST00000258042.2
|
NMBR
|
neuromedin B receptor |
| chr1_-_203086001 | 0.14 |
ENST00000241651.5
|
MYOG
|
myogenin |
| chr12_+_7189675 | 0.14 |
ENST00000675855.1
ENST00000434354.6 ENST00000544456.5 ENST00000545574.5 ENST00000420616.6 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr2_+_134254065 | 0.14 |
ENST00000281923.4
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase |
| chr6_-_109440504 | 0.13 |
ENST00000520723.5
ENST00000518648.1 ENST00000417394.6 ENST00000521072.7 |
PPIL6
|
peptidylprolyl isomerase like 6 |
| chr7_-_100656384 | 0.13 |
ENST00000461605.1
ENST00000160382.10 |
ACTL6B
|
actin like 6B |
| chr19_+_48393657 | 0.13 |
ENST00000263269.4
|
GRIN2D
|
glutamate ionotropic receptor NMDA type subunit 2D |
| chr12_+_50057548 | 0.13 |
ENST00000228468.8
ENST00000447966.7 |
ASIC1
|
acid sensing ion channel subunit 1 |
| chr22_-_37427433 | 0.12 |
ENST00000452946.1
ENST00000402918.7 |
ELFN2
ELFN2
|
extracellular leucine rich repeat and fibronectin type III domain containing 2 extracellular leucine rich repeat and fibronectin type III domain containing 2 |
| chr6_-_35512863 | 0.12 |
ENST00000428978.1
ENST00000614066.4 ENST00000322263.8 |
TULP1
|
TUB like protein 1 |
| chr20_+_31475278 | 0.12 |
ENST00000201979.3
|
REM1
|
RRAD and GEM like GTPase 1 |
| chr19_-_42242526 | 0.11 |
ENST00000222330.8
ENST00000676537.1 |
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr4_-_165112852 | 0.11 |
ENST00000505095.1
ENST00000306480.11 |
TMEM192
|
transmembrane protein 192 |
| chr19_-_35900532 | 0.11 |
ENST00000396901.5
ENST00000641389.2 ENST00000585925.7 |
NFKBID
|
NFKB inhibitor delta |
| chr14_-_93115869 | 0.11 |
ENST00000556603.6
ENST00000354313.7 ENST00000267615.11 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr12_-_57129001 | 0.10 |
ENST00000556155.5
|
STAT6
|
signal transducer and activator of transcription 6 |
| chr11_-_61891381 | 0.10 |
ENST00000525588.5
|
FADS3
|
fatty acid desaturase 3 |
| chr14_-_93115812 | 0.10 |
ENST00000553452.5
|
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr8_+_30384511 | 0.10 |
ENST00000339877.8
ENST00000320203.8 ENST00000287771.9 ENST00000397323.9 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr15_-_41230697 | 0.10 |
ENST00000314992.9
ENST00000558396.1 ENST00000458580.7 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr20_+_23035312 | 0.10 |
ENST00000255008.5
|
SSTR4
|
somatostatin receptor 4 |
| chr19_-_12881460 | 0.10 |
ENST00000592506.1
|
DNASE2
|
deoxyribonuclease 2, lysosomal |
| chr10_-_75099489 | 0.10 |
ENST00000472493.6
ENST00000478873.7 ENST00000605915.5 |
DUSP13
|
dual specificity phosphatase 13 |
| chr17_+_79024142 | 0.10 |
ENST00000579760.6
ENST00000339142.6 |
C1QTNF1
|
C1q and TNF related 1 |
| chr19_-_4066892 | 0.09 |
ENST00000322357.9
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr2_+_62705644 | 0.09 |
ENST00000427809.5
ENST00000405482.5 ENST00000431489.6 |
EHBP1
|
EH domain binding protein 1 |
| chr17_+_79024243 | 0.09 |
ENST00000311661.4
|
C1QTNF1
|
C1q and TNF related 1 |
| chr19_-_11481044 | 0.09 |
ENST00000359227.8
|
ELAVL3
|
ELAV like RNA binding protein 3 |
| chr4_-_119628007 | 0.09 |
ENST00000420633.1
ENST00000394439.5 |
PDE5A
|
phosphodiesterase 5A |
| chr7_-_103989649 | 0.09 |
ENST00000428762.6
|
RELN
|
reelin |
| chr10_-_119536533 | 0.09 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr9_-_128724088 | 0.09 |
ENST00000406904.2
ENST00000452105.5 ENST00000372667.9 ENST00000372663.9 |
ZDHHC12
|
zinc finger DHHC-type palmitoyltransferase 12 |
| chr6_-_35512882 | 0.08 |
ENST00000229771.11
|
TULP1
|
TUB like protein 1 |
| chr6_+_11093753 | 0.08 |
ENST00000416247.4
|
SMIM13
|
small integral membrane protein 13 |
| chr19_-_49867251 | 0.07 |
ENST00000631020.2
ENST00000596014.5 ENST00000636994.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr19_+_41264345 | 0.07 |
ENST00000378215.8
ENST00000392006.8 ENST00000617774.1 ENST00000602130.5 ENST00000617305.4 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U like 1 |
| chr3_+_122795039 | 0.07 |
ENST00000261038.6
|
SLC49A4
|
solute carrier family 49 member 4 |
| chr21_-_17612842 | 0.07 |
ENST00000339775.10
ENST00000348354.7 |
BTG3
|
BTG anti-proliferation factor 3 |
| chr1_+_1512137 | 0.07 |
ENST00000378756.8
ENST00000378755.9 |
ATAD3A
|
ATPase family AAA domain containing 3A |
| chr16_+_71358713 | 0.06 |
ENST00000349553.9
ENST00000302628.9 ENST00000562305.5 |
CALB2
|
calbindin 2 |
| chr10_+_112950240 | 0.06 |
ENST00000627217.3
ENST00000534894.5 ENST00000538897.5 ENST00000536810.5 ENST00000355995.8 ENST00000542695.5 ENST00000543371.5 |
TCF7L2
|
transcription factor 7 like 2 |
| chr19_+_4279285 | 0.06 |
ENST00000599689.1
|
SHD
|
Src homology 2 domain containing transforming protein D |
| chr9_-_16870662 | 0.06 |
ENST00000380672.9
|
BNC2
|
basonuclin 2 |
| chr1_-_101996919 | 0.06 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chr3_-_52897541 | 0.06 |
ENST00000355083.11
ENST00000504329.1 |
STIMATE
STIMATE-MUSTN1
|
STIM activating enhancer STIMATE-MUSTN1 readthrough |
| chr5_+_151020438 | 0.06 |
ENST00000622181.4
ENST00000614343.4 ENST00000388825.9 ENST00000521650.5 ENST00000517973.1 |
GPX3
|
glutathione peroxidase 3 |
| chr9_-_37904085 | 0.06 |
ENST00000377716.6
ENST00000242275.7 |
SLC25A51
|
solute carrier family 25 member 51 |
| chr3_+_184338826 | 0.06 |
ENST00000453072.5
|
FAM131A
|
family with sequence similarity 131 member A |
| chr19_-_55599493 | 0.05 |
ENST00000221665.5
ENST00000592585.1 |
FIZ1
|
FLT3 interacting zinc finger 1 |
| chr12_-_118372883 | 0.05 |
ENST00000542532.5
ENST00000392533.8 |
TAOK3
|
TAO kinase 3 |
| chr11_-_30586866 | 0.05 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr19_+_45079195 | 0.05 |
ENST00000591607.1
ENST00000591747.5 ENST00000270257.9 ENST00000391951.2 ENST00000587566.5 |
GEMIN7
MARK4
|
gem nuclear organelle associated protein 7 microtubule affinity regulating kinase 4 |
| chr17_-_81239025 | 0.05 |
ENST00000637944.2
|
TEPSIN
|
TEPSIN adaptor related protein complex 4 accessory protein |
| chr8_+_22165358 | 0.05 |
ENST00000306349.13
ENST00000306385.10 |
BMP1
|
bone morphogenetic protein 1 |
| chr11_-_107858777 | 0.05 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr19_+_4279247 | 0.05 |
ENST00000543264.7
|
SHD
|
Src homology 2 domain containing transforming protein D |
| chr11_-_30586344 | 0.05 |
ENST00000358117.10
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chrX_+_137566119 | 0.05 |
ENST00000287538.10
|
ZIC3
|
Zic family member 3 |
| chr1_+_53062052 | 0.05 |
ENST00000395871.7
ENST00000673702.1 ENST00000673956.1 ENST00000312553.10 ENST00000371500.8 ENST00000618387.1 |
PODN
|
podocan |
| chr1_+_1471751 | 0.04 |
ENST00000673477.1
ENST00000308647.8 |
ATAD3B
|
ATPase family AAA domain containing 3B |
| chr11_-_1036706 | 0.04 |
ENST00000421673.7
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr4_-_20984011 | 0.04 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr12_-_49707220 | 0.04 |
ENST00000550488.5
|
FMNL3
|
formin like 3 |
| chr7_-_27180230 | 0.04 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr1_+_26317950 | 0.03 |
ENST00000374213.3
|
CD52
|
CD52 molecule |
| chr15_-_58933668 | 0.03 |
ENST00000380516.7
|
SLTM
|
SAFB like transcription modulator |
| chr7_+_151051201 | 0.03 |
ENST00000490540.1
|
ASIC3
|
acid sensing ion channel subunit 3 |
| chr11_-_61891534 | 0.03 |
ENST00000278829.7
|
FADS3
|
fatty acid desaturase 3 |
| chr17_-_44376169 | 0.03 |
ENST00000587295.5
|
ITGA2B
|
integrin subunit alpha 2b |
| chr6_+_42929430 | 0.03 |
ENST00000372836.5
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr9_-_96619783 | 0.03 |
ENST00000375241.6
|
CDC14B
|
cell division cycle 14B |
| chr12_+_52079700 | 0.02 |
ENST00000546390.2
|
SMIM41
|
small integral membrane protein 41 |
| chr6_+_142147162 | 0.02 |
ENST00000452973.6
ENST00000620996.4 ENST00000367621.1 ENST00000367630.9 |
VTA1
|
vesicle trafficking 1 |
| chr7_+_29194757 | 0.02 |
ENST00000222792.11
|
CHN2
|
chimerin 2 |
| chr1_-_204152010 | 0.02 |
ENST00000367202.9
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_204151884 | 0.02 |
ENST00000367201.7
|
ETNK2
|
ethanolamine kinase 2 |
| chr11_-_63015831 | 0.02 |
ENST00000430500.6
ENST00000336232.7 |
SLC22A8
|
solute carrier family 22 member 8 |
| chr16_+_70114306 | 0.02 |
ENST00000288050.9
ENST00000398122.7 ENST00000568530.5 |
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit |
| chr3_+_32685128 | 0.02 |
ENST00000331889.10
ENST00000328834.9 |
CNOT10
|
CCR4-NOT transcription complex subunit 10 |
| chr22_+_35257452 | 0.02 |
ENST00000420166.5
ENST00000216106.6 ENST00000455359.5 |
HMGXB4
|
HMG-box containing 4 |
| chr11_-_63917129 | 0.02 |
ENST00000301459.5
|
RCOR2
|
REST corepressor 2 |
| chr1_+_26993684 | 0.02 |
ENST00000522111.3
|
TRNP1
|
TMF1 regulated nuclear protein 1 |
| chr3_-_69080350 | 0.02 |
ENST00000630585.1
ENST00000361055.9 ENST00000415609.6 ENST00000349511.8 |
UBA3
|
ubiquitin like modifier activating enzyme 3 |
| chr13_-_46052712 | 0.01 |
ENST00000242848.8
ENST00000679008.1 ENST00000282007.7 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr9_-_127980976 | 0.01 |
ENST00000373095.6
|
FAM102A
|
family with sequence similarity 102 member A |
| chr9_-_96619378 | 0.01 |
ENST00000375240.7
ENST00000463569.5 |
CDC14B
|
cell division cycle 14B |
| chr2_-_206086057 | 0.01 |
ENST00000403263.6
|
INO80D
|
INO80 complex subunit D |
| chr19_+_35533436 | 0.01 |
ENST00000222286.9
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr19_-_46714269 | 0.01 |
ENST00000600194.5
|
PRKD2
|
protein kinase D2 |
| chr6_+_42929127 | 0.01 |
ENST00000394142.7
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr16_+_283157 | 0.01 |
ENST00000219406.11
ENST00000404312.5 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A member 2 |
| chr18_-_5543988 | 0.01 |
ENST00000341928.7
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr8_+_22165140 | 0.01 |
ENST00000397814.7
ENST00000354870.5 |
BMP1
|
bone morphogenetic protein 1 |
| chr2_+_14632688 | 0.01 |
ENST00000331243.4
ENST00000295092.3 |
LRATD1
|
LRAT domain containing 1 |
| chr19_-_15200902 | 0.01 |
ENST00000601011.1
ENST00000263388.7 |
NOTCH3
|
notch receptor 3 |
| chr2_+_170929198 | 0.00 |
ENST00000234160.5
|
GORASP2
|
golgi reassembly stacking protein 2 |
| chr19_-_56671746 | 0.00 |
ENST00000537055.4
ENST00000601659.1 |
ZNF835
|
zinc finger protein 835 |
| chr12_+_57128656 | 0.00 |
ENST00000338962.8
|
LRP1
|
LDL receptor related protein 1 |
| chr9_-_127897399 | 0.00 |
ENST00000542456.5
ENST00000373141.5 |
ST6GALNAC6
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF423
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0036304 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.2 | 0.5 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.5 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.4 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:1990834 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.1 | 1.0 | GO:0051798 | negative regulation of activin receptor signaling pathway(GO:0032926) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.8 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.2 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.1 | 0.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |