Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
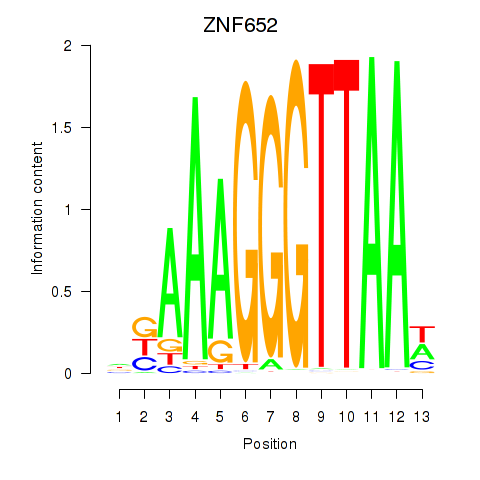
Results for ZNF652
Z-value: 1.65
Transcription factors associated with ZNF652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF652
|
ENSG00000198740.9 | zinc finger protein 652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF652 | hg38_v1_chr17_-_49362206_49362279 | 0.58 | 1.3e-01 | Click! |
Activity profile of ZNF652 motif
Sorted Z-values of ZNF652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_72216745 | 3.62 |
ENST00000517827.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr3_+_157436842 | 2.49 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr11_-_35525603 | 2.35 |
ENST00000529303.1
ENST00000619888.5 ENST00000622144.4 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr11_-_35526024 | 2.29 |
ENST00000615849.4
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr2_-_187513641 | 2.10 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr11_-_63614425 | 2.05 |
ENST00000415826.3
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr2_-_189179754 | 1.99 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr13_-_37598750 | 1.94 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr17_-_19386785 | 1.91 |
ENST00000497081.6
|
MFAP4
|
microfibril associated protein 4 |
| chr17_-_19387170 | 1.75 |
ENST00000395592.6
ENST00000299610.5 |
MFAP4
|
microfibril associated protein 4 |
| chr5_-_76623391 | 1.75 |
ENST00000296641.5
ENST00000504899.1 |
F2RL2
|
coagulation factor II thrombin receptor like 2 |
| chr3_-_112641128 | 1.74 |
ENST00000206423.8
|
CCDC80
|
coiled-coil domain containing 80 |
| chr6_+_72216442 | 1.72 |
ENST00000425662.6
ENST00000453976.6 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr1_+_215082731 | 1.71 |
ENST00000444842.7
|
KCNK2
|
potassium two pore domain channel subfamily K member 2 |
| chrX_-_155022716 | 1.68 |
ENST00000360256.9
|
F8
|
coagulation factor VIII |
| chr22_-_24245059 | 1.66 |
ENST00000398292.3
ENST00000263112.11 ENST00000327365.10 ENST00000424217.1 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr3_-_112641292 | 1.49 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr6_-_52577012 | 1.42 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr17_+_50095285 | 1.38 |
ENST00000503614.5
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr9_+_117704168 | 1.34 |
ENST00000472304.2
ENST00000394487.5 |
TLR4
|
toll like receptor 4 |
| chr3_+_148739798 | 1.30 |
ENST00000402260.2
|
AGTR1
|
angiotensin II receptor type 1 |
| chr2_-_68319887 | 1.30 |
ENST00000409862.1
ENST00000263655.4 |
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr2_+_151357583 | 1.24 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr3_-_9952337 | 1.23 |
ENST00000411976.2
ENST00000412055.6 |
PRRT3
|
proline rich transmembrane protein 3 |
| chr17_+_50095331 | 1.17 |
ENST00000503176.6
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr1_+_197902720 | 1.09 |
ENST00000436652.1
|
C1orf53
|
chromosome 1 open reading frame 53 |
| chr11_-_111911759 | 1.08 |
ENST00000650687.2
|
CRYAB
|
crystallin alpha B |
| chr2_+_171522466 | 1.07 |
ENST00000321348.9
ENST00000375252.3 |
CYBRD1
|
cytochrome b reductase 1 |
| chr10_-_28334439 | 1.07 |
ENST00000441595.2
|
MPP7
|
membrane palmitoylated protein 7 |
| chrX_+_103776831 | 1.04 |
ENST00000621218.5
ENST00000619236.1 |
PLP1
|
proteolipid protein 1 |
| chr9_-_121050264 | 1.04 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr6_-_31729260 | 1.02 |
ENST00000375789.7
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_+_201739864 | 1.02 |
ENST00000367295.5
|
NAV1
|
neuron navigator 1 |
| chr16_-_3305397 | 1.01 |
ENST00000396862.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr16_+_53208438 | 1.00 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_31729478 | 1.00 |
ENST00000436437.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr2_+_171522227 | 0.98 |
ENST00000409484.5
|
CYBRD1
|
cytochrome b reductase 1 |
| chr21_-_26843063 | 0.96 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843012 | 0.94 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr3_+_97439603 | 0.93 |
ENST00000514100.5
|
EPHA6
|
EPH receptor A6 |
| chr6_-_31729785 | 0.92 |
ENST00000416410.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_+_76681208 | 0.88 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr11_-_119423162 | 0.88 |
ENST00000284240.10
ENST00000524970.5 |
THY1
|
Thy-1 cell surface antigen |
| chr1_+_197902607 | 0.86 |
ENST00000367393.8
|
C1orf53
|
chromosome 1 open reading frame 53 |
| chr16_+_31472130 | 0.85 |
ENST00000394863.8
ENST00000565360.5 ENST00000361773.7 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr10_-_13972355 | 0.83 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr6_-_31730198 | 0.81 |
ENST00000375787.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_+_75605462 | 0.79 |
ENST00000539311.5
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr1_-_32702736 | 0.78 |
ENST00000373484.4
ENST00000409190.8 |
SYNC
|
syncoilin, intermediate filament protein |
| chr3_-_15797930 | 0.77 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr2_-_144517663 | 0.76 |
ENST00000427902.5
ENST00000462355.2 ENST00000470879.5 ENST00000409487.7 ENST00000435831.5 ENST00000630572.2 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_+_51861604 | 0.75 |
ENST00000333127.4
|
IQCF2
|
IQ motif containing F2 |
| chr1_-_182672232 | 0.74 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr1_+_197912462 | 0.73 |
ENST00000475727.1
ENST00000367391.5 ENST00000367390.7 |
LHX9
|
LIM homeobox 9 |
| chr12_+_6873561 | 0.73 |
ENST00000433346.5
|
LRRC23
|
leucine rich repeat containing 23 |
| chr5_+_140125935 | 0.71 |
ENST00000333305.5
|
IGIP
|
IgA inducing protein |
| chr17_-_65561640 | 0.71 |
ENST00000618960.4
ENST00000307078.10 |
AXIN2
|
axin 2 |
| chr10_-_13707536 | 0.69 |
ENST00000632570.1
ENST00000477221.2 |
FRMD4A
|
FERM domain containing 4A |
| chr11_+_57460493 | 0.67 |
ENST00000335099.8
|
RTN4RL2
|
reticulon 4 receptor like 2 |
| chr4_+_128809791 | 0.64 |
ENST00000452328.6
ENST00000504089.5 |
JADE1
|
jade family PHD finger 1 |
| chr15_+_63277586 | 0.61 |
ENST00000261879.10
ENST00000380343.8 ENST00000560353.1 |
APH1B
|
aph-1 homolog B, gamma-secretase subunit |
| chr1_-_156677400 | 0.61 |
ENST00000368223.4
|
NES
|
nestin |
| chr7_+_80602200 | 0.61 |
ENST00000534394.5
|
CD36
|
CD36 molecule |
| chr2_-_73233206 | 0.59 |
ENST00000258083.3
|
PRADC1
|
protease associated domain containing 1 |
| chr7_+_80602150 | 0.59 |
ENST00000309881.11
|
CD36
|
CD36 molecule |
| chr5_-_178627001 | 0.57 |
ENST00000611575.4
ENST00000520957.1 ENST00000316308.9 ENST00000611733.4 |
CLK4
|
CDC like kinase 4 |
| chr14_-_100568070 | 0.57 |
ENST00000557378.6
ENST00000443071.6 ENST00000637646.1 |
BEGAIN
|
brain enriched guanylate kinase associated |
| chr4_+_128809684 | 0.56 |
ENST00000226319.11
ENST00000511647.5 |
JADE1
|
jade family PHD finger 1 |
| chr11_-_63614366 | 0.55 |
ENST00000323646.9
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr8_+_1973668 | 0.55 |
ENST00000320248.4
|
KBTBD11
|
kelch repeat and BTB domain containing 11 |
| chr16_+_3305472 | 0.55 |
ENST00000574298.6
ENST00000669516.2 |
ZNF75A
|
zinc finger protein 75a |
| chr11_+_45146631 | 0.54 |
ENST00000534751.3
ENST00000683152.1 |
PRDM11
|
PR/SET domain 11 |
| chr14_-_100568475 | 0.54 |
ENST00000553553.6
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr11_-_86672114 | 0.53 |
ENST00000393324.7
|
ME3
|
malic enzyme 3 |
| chr17_+_45148468 | 0.52 |
ENST00000332499.4
|
HEXIM1
|
HEXIM P-TEFb complex subunit 1 |
| chr15_+_67128103 | 0.52 |
ENST00000558894.5
|
SMAD3
|
SMAD family member 3 |
| chr4_+_70242583 | 0.52 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr11_-_86672419 | 0.51 |
ENST00000524826.7
ENST00000532471.1 |
ME3
|
malic enzyme 3 |
| chr1_+_149475760 | 0.51 |
ENST00000611931.4
|
NBPF19
|
NBPF member 19 |
| chr7_-_138627444 | 0.50 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr1_+_22025487 | 0.50 |
ENST00000634934.2
ENST00000634712.2 ENST00000634451.2 ENST00000635450.2 ENST00000420503.1 ENST00000416769.2 ENST00000642072.1 ENST00000404210.5 ENST00000641009.1 ENST00000648594.1 |
LINC00339
CDC42
|
long intergenic non-protein coding RNA 339 cell division cycle 42 |
| chr19_-_45782388 | 0.50 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr2_+_157257687 | 0.50 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr7_+_12686849 | 0.50 |
ENST00000396662.5
ENST00000356797.7 ENST00000396664.2 |
ARL4A
|
ADP ribosylation factor like GTPase 4A |
| chr12_+_56152439 | 0.50 |
ENST00000550443.5
|
MYL6B
|
myosin light chain 6B |
| chrX_-_101052054 | 0.50 |
ENST00000372939.5
ENST00000372935.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B |
| chr15_-_63157464 | 0.49 |
ENST00000330964.10
ENST00000635699.1 ENST00000439025.1 |
RPS27L
|
ribosomal protein S27 like |
| chrX_-_101052087 | 0.49 |
ENST00000372936.4
ENST00000545398.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B |
| chr7_+_12687625 | 0.49 |
ENST00000651779.1
ENST00000404894.1 |
ARL4A
|
ADP ribosylation factor like GTPase 4A |
| chr12_+_56152579 | 0.47 |
ENST00000551834.5
ENST00000552568.5 |
MYL6B
|
myosin light chain 6B |
| chr12_+_75480800 | 0.46 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr1_-_150808251 | 0.45 |
ENST00000271651.8
ENST00000676970.1 ENST00000679260.1 ENST00000676751.1 ENST00000677887.1 |
CTSK
|
cathepsin K |
| chr2_+_161160299 | 0.44 |
ENST00000440506.5
ENST00000429217.5 ENST00000406287.5 ENST00000402568.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr17_+_45999250 | 0.44 |
ENST00000537309.1
|
STH
|
saitohin |
| chr17_+_28473635 | 0.43 |
ENST00000314669.10
ENST00000545060.2 |
SLC13A2
|
solute carrier family 13 member 2 |
| chr16_-_66550091 | 0.43 |
ENST00000564917.5
ENST00000677420.1 |
TK2
|
thymidine kinase 2 |
| chr19_+_17215382 | 0.43 |
ENST00000595101.5
ENST00000596136.5 ENST00000445667.6 |
USE1
|
unconventional SNARE in the ER 1 |
| chr3_-_142000353 | 0.43 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr2_+_201129318 | 0.41 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr16_-_66549839 | 0.41 |
ENST00000527800.6
ENST00000677555.1 ENST00000563369.6 |
TK2
|
thymidine kinase 2 |
| chr3_-_179259208 | 0.41 |
ENST00000485523.5
|
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr6_+_39792993 | 0.40 |
ENST00000538976.5
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr17_+_28473278 | 0.40 |
ENST00000444914.7
|
SLC13A2
|
solute carrier family 13 member 2 |
| chr1_+_61082702 | 0.40 |
ENST00000485903.6
ENST00000371185.6 ENST00000371184.6 |
NFIA
|
nuclear factor I A |
| chr6_+_39793008 | 0.39 |
ENST00000398904.6
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr1_-_182671902 | 0.39 |
ENST00000483095.6
|
RGS8
|
regulator of G protein signaling 8 |
| chr19_+_39412650 | 0.39 |
ENST00000425673.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr1_-_182671853 | 0.39 |
ENST00000367556.5
|
RGS8
|
regulator of G protein signaling 8 |
| chrX_-_135296024 | 0.39 |
ENST00000370764.1
|
ZNF75D
|
zinc finger protein 75D |
| chr16_-_30787169 | 0.39 |
ENST00000262525.6
|
ZNF629
|
zinc finger protein 629 |
| chr16_-_79600698 | 0.38 |
ENST00000393350.1
|
MAF
|
MAF bZIP transcription factor |
| chr11_-_89491131 | 0.37 |
ENST00000343727.9
ENST00000531342.5 ENST00000375979.7 |
NOX4
|
NADPH oxidase 4 |
| chr9_+_128922283 | 0.36 |
ENST00000421063.6
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr1_+_149475933 | 0.36 |
ENST00000612881.4
|
NBPF19
|
NBPF member 19 |
| chr16_-_66550112 | 0.36 |
ENST00000544898.6
ENST00000620035.5 ENST00000545043.6 |
TK2
|
thymidine kinase 2 |
| chr16_-_66550005 | 0.36 |
ENST00000527284.6
|
TK2
|
thymidine kinase 2 |
| chr2_+_203328378 | 0.35 |
ENST00000430418.5
ENST00000261018.12 ENST00000295851.10 ENST00000424558.5 ENST00000417864.5 ENST00000422511.6 |
ABI2
|
abl interactor 2 |
| chr9_+_128411715 | 0.35 |
ENST00000420034.5
ENST00000372842.5 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr3_-_48635426 | 0.35 |
ENST00000455886.6
ENST00000431739.5 ENST00000426599.1 ENST00000383733.7 ENST00000395550.7 ENST00000420764.6 ENST00000337000.12 |
SLC26A6
|
solute carrier family 26 member 6 |
| chr2_-_98608452 | 0.35 |
ENST00000328709.8
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr2_+_203328266 | 0.35 |
ENST00000261017.9
|
ABI2
|
abl interactor 2 |
| chr10_-_28282086 | 0.35 |
ENST00000375719.7
ENST00000375732.5 |
MPP7
|
membrane palmitoylated protein 7 |
| chr7_-_150978284 | 0.35 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2 |
| chr6_+_125781108 | 0.34 |
ENST00000368357.7
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr16_+_53207981 | 0.34 |
ENST00000565803.2
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr3_-_15798184 | 0.34 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr4_-_98657635 | 0.34 |
ENST00000515287.5
ENST00000511651.5 ENST00000505184.5 |
TSPAN5
|
tetraspanin 5 |
| chr3_-_127722562 | 0.33 |
ENST00000487473.5
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chr19_-_45782479 | 0.33 |
ENST00000447742.6
ENST00000354227.9 ENST00000291270.9 ENST00000683086.1 ENST00000343373.10 |
DMPK
|
DM1 protein kinase |
| chrX_+_100666854 | 0.33 |
ENST00000640282.1
|
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr17_-_81166160 | 0.33 |
ENST00000326724.9
|
AATK
|
apoptosis associated tyrosine kinase |
| chr7_-_138002017 | 0.33 |
ENST00000452463.5
ENST00000456390.5 ENST00000330387.11 |
CREB3L2
|
cAMP responsive element binding protein 3 like 2 |
| chr4_-_76213724 | 0.33 |
ENST00000639738.1
|
SCARB2
|
scavenger receptor class B member 2 |
| chr19_+_41003946 | 0.32 |
ENST00000593831.1
|
CYP2B6
|
cytochrome P450 family 2 subfamily B member 6 |
| chr12_-_6700788 | 0.32 |
ENST00000320591.9
ENST00000534837.6 |
PIANP
|
PILR alpha associated neural protein |
| chr16_-_79600727 | 0.32 |
ENST00000326043.5
|
MAF
|
MAF bZIP transcription factor |
| chr19_+_10420474 | 0.31 |
ENST00000380702.7
|
PDE4A
|
phosphodiesterase 4A |
| chr2_+_161160420 | 0.31 |
ENST00000392749.7
ENST00000405852.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr1_-_146144804 | 0.31 |
ENST00000583866.9
ENST00000617010.2 |
NBPF10
|
NBPF member 10 |
| chr12_-_6873278 | 0.31 |
ENST00000523102.5
ENST00000524270.6 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr3_+_111674654 | 0.31 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr12_+_120446418 | 0.31 |
ENST00000551765.6
ENST00000229384.5 |
GATC
|
glutamyl-tRNA amidotransferase subunit C |
| chr14_+_58637934 | 0.30 |
ENST00000395153.8
|
DACT1
|
dishevelled binding antagonist of beta catenin 1 |
| chr12_+_103587266 | 0.30 |
ENST00000388887.7
|
STAB2
|
stabilin 2 |
| chr4_-_7871986 | 0.30 |
ENST00000360265.9
|
AFAP1
|
actin filament associated protein 1 |
| chr20_-_3173516 | 0.30 |
ENST00000360342.7
ENST00000645462.1 ENST00000337576.7 |
LZTS3
|
leucine zipper tumor suppressor family member 3 |
| chr7_-_138001794 | 0.30 |
ENST00000616381.4
ENST00000620715.4 |
CREB3L2
|
cAMP responsive element binding protein 3 like 2 |
| chr20_-_45124463 | 0.30 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr8_-_27772585 | 0.30 |
ENST00000522915.5
ENST00000356537.9 |
CCDC25
|
coiled-coil domain containing 25 |
| chr3_+_138621225 | 0.30 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_-_23302823 | 0.30 |
ENST00000452015.9
|
PPP1R3E
|
protein phosphatase 1 regulatory subunit 3E |
| chr4_-_76213589 | 0.29 |
ENST00000638603.1
ENST00000452464.6 |
SCARB2
|
scavenger receptor class B member 2 |
| chr3_-_64445396 | 0.29 |
ENST00000295902.11
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr19_-_46634685 | 0.29 |
ENST00000300873.4
|
GNG8
|
G protein subunit gamma 8 |
| chr4_-_185810894 | 0.29 |
ENST00000448662.6
ENST00000439049.5 ENST00000420158.5 ENST00000319471.13 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_56557155 | 0.28 |
ENST00000422222.5
ENST00000394672.8 ENST00000326595.11 |
CCDC66
|
coiled-coil domain containing 66 |
| chr16_-_66550142 | 0.27 |
ENST00000417693.8
ENST00000299697.12 ENST00000451102.7 |
TK2
|
thymidine kinase 2 |
| chr9_-_35815034 | 0.27 |
ENST00000259667.6
|
HINT2
|
histidine triad nucleotide binding protein 2 |
| chr14_-_57866075 | 0.27 |
ENST00000556826.6
|
SLC35F4
|
solute carrier family 35 member F4 |
| chr6_-_28252246 | 0.26 |
ENST00000377294.3
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr1_-_159925496 | 0.26 |
ENST00000368097.9
|
TAGLN2
|
transgelin 2 |
| chr8_-_109644766 | 0.26 |
ENST00000533065.5
ENST00000276646.14 |
SYBU
|
syntabulin |
| chr2_+_161136901 | 0.26 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr17_+_7425606 | 0.26 |
ENST00000333870.8
ENST00000574034.1 |
SPEM2
|
SPEM family member 2 |
| chr15_-_56918571 | 0.26 |
ENST00000559000.6
|
ENSG00000285253.1
|
novel protein |
| chr17_-_44830774 | 0.26 |
ENST00000590758.3
ENST00000591424.5 |
GJC1
|
gap junction protein gamma 1 |
| chr3_+_53494591 | 0.26 |
ENST00000288139.11
ENST00000350061.11 ENST00000636938.1 |
CACNA1D
|
calcium voltage-gated channel subunit alpha1 D |
| chr9_-_113221288 | 0.26 |
ENST00000446284.6
ENST00000414250.2 |
FKBP15
|
FKBP prolyl isomerase family member 15 |
| chr1_+_61082398 | 0.25 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A |
| chr5_+_42548043 | 0.25 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr2_+_201129483 | 0.25 |
ENST00000440180.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr16_-_1793694 | 0.25 |
ENST00000569769.1
ENST00000215539.4 |
SPSB3
IGFALS
|
splA/ryanodine receptor domain and SOCS box containing 3 insulin like growth factor binding protein acid labile subunit |
| chr1_-_1600081 | 0.24 |
ENST00000422725.4
|
FNDC10
|
fibronectin type III domain containing 10 |
| chrX_+_52897306 | 0.24 |
ENST00000612188.4
ENST00000616419.4 ENST00000416841.7 ENST00000593751.7 |
FAM156B
|
family with sequence similarity 156 member B |
| chr16_-_1793719 | 0.24 |
ENST00000415638.3
|
IGFALS
|
insulin like growth factor binding protein acid labile subunit |
| chr21_+_39445824 | 0.24 |
ENST00000380637.7
ENST00000380634.5 ENST00000458295.5 ENST00000440288.6 ENST00000380631.5 |
SH3BGR
|
SH3 domain binding glutamate rich protein |
| chr5_-_157575767 | 0.24 |
ENST00000257527.9
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr17_+_45241067 | 0.24 |
ENST00000587489.5
|
FMNL1
|
formin like 1 |
| chr14_+_79279403 | 0.24 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr14_+_79279339 | 0.24 |
ENST00000557594.5
|
NRXN3
|
neurexin 3 |
| chr10_+_5048748 | 0.24 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr5_+_141364231 | 0.24 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr2_-_15561305 | 0.23 |
ENST00000281513.10
|
NBAS
|
NBAS subunit of NRZ tethering complex |
| chr13_+_114314474 | 0.23 |
ENST00000463003.2
ENST00000645174.1 ENST00000361283.4 ENST00000644294.1 |
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1 |
| chr10_+_122560679 | 0.23 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr17_+_39688079 | 0.23 |
ENST00000578199.5
|
ERBB2
|
erb-b2 receptor tyrosine kinase 2 |
| chr7_+_102285125 | 0.23 |
ENST00000536178.3
|
SH2B2
|
SH2B adaptor protein 2 |
| chr19_+_17215332 | 0.23 |
ENST00000263897.10
|
USE1
|
unconventional SNARE in the ER 1 |
| chr4_-_164977644 | 0.23 |
ENST00000329314.6
ENST00000508856.2 |
TRIM61
|
tripartite motif containing 61 |
| chr5_-_78985288 | 0.23 |
ENST00000264914.10
|
ARSB
|
arylsulfatase B |
| chr1_+_100719734 | 0.22 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr3_+_138621207 | 0.22 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_+_79279681 | 0.22 |
ENST00000679122.1
|
NRXN3
|
neurexin 3 |
| chr16_+_4624811 | 0.22 |
ENST00000415496.5
ENST00000262370.12 ENST00000587747.5 ENST00000399577.9 ENST00000588994.5 ENST00000586183.5 |
MGRN1
|
mahogunin ring finger 1 |
| chr11_-_66336396 | 0.22 |
ENST00000627248.1
ENST00000311320.9 |
RIN1
|
Ras and Rab interactor 1 |
| chr5_-_115602416 | 0.22 |
ENST00000427199.3
|
TICAM2
|
toll like receptor adaptor molecule 2 |
| chr17_-_4786433 | 0.22 |
ENST00000354194.4
|
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr14_+_24120956 | 0.21 |
ENST00000558325.2
|
ENSG00000259371.2
|
novel protein |
| chr10_+_122560751 | 0.21 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr6_-_131000722 | 0.21 |
ENST00000528282.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF652
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.7 | 2.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 1.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.4 | 1.3 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.3 | 1.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 3.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.8 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.3 | 0.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 0.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 2.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 5.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 1.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 3.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 1.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.5 | GO:0071338 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.1 | 1.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 2.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 1.2 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 1.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.7 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.7 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 0.2 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 1.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 1.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.7 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.4 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.1 | 0.2 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 0.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.2 | GO:0051714 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.3 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:2000538 | positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 1.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 1.0 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 1.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.7 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 2.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:1904778 | protein localization to cell cortex(GO:0072697) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 1.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.8 | GO:0001510 | RNA methylation(GO:0001510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 2.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.4 | 3.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 3.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 4.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.2 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 1.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.2 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.1 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 2.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 3.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.4 | 1.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.4 | 2.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 2.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.3 | 2.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.8 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.3 | 1.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.0 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 1.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.2 | 1.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 0.8 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 1.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 3.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.4 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.2 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 1.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 4.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 4.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 7.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 3.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 2.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 2.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.4 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |