Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
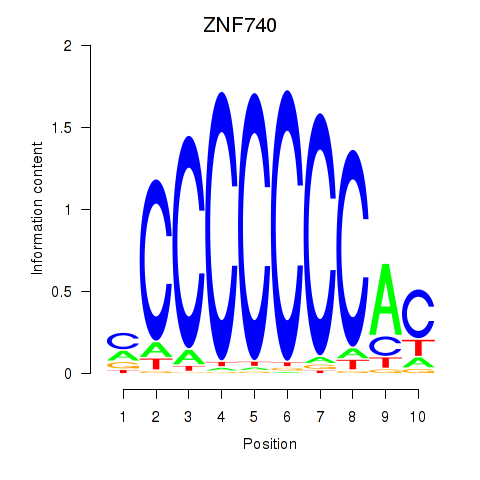
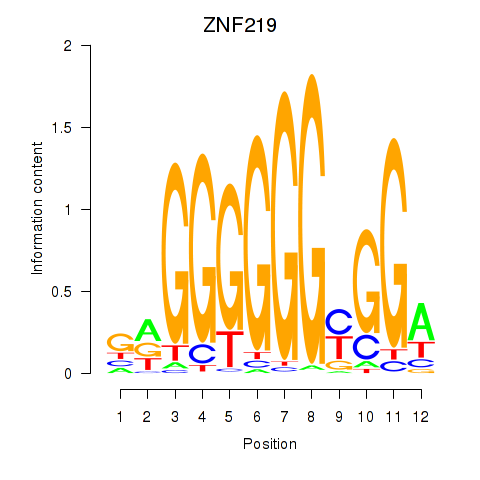
Results for ZNF740_ZNF219
Z-value: 0.82
Transcription factors associated with ZNF740_ZNF219
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF740
|
ENSG00000139651.11 | zinc finger protein 740 |
|
ZNF219
|
ENSG00000165804.16 | zinc finger protein 219 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF219 | hg38_v1_chr14_-_21094488_21094500 | 0.96 | 1.4e-04 | Click! |
| ZNF740 | hg38_v1_chr12_+_53180679_53180755 | 0.96 | 1.5e-04 | Click! |
Activity profile of ZNF740_ZNF219 motif
Sorted Z-values of ZNF740_ZNF219 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94641145 | 1.68 |
ENST00000433389.8
ENST00000358397.9 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr20_+_59604527 | 1.67 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr8_+_94641074 | 1.65 |
ENST00000423620.6
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr8_+_94641199 | 1.64 |
ENST00000646773.1
ENST00000454170.7 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr11_-_64166102 | 0.80 |
ENST00000255681.7
ENST00000675777.1 |
MACROD1
|
mono-ADP ribosylhydrolase 1 |
| chr17_+_7884783 | 0.74 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr14_-_105168753 | 0.61 |
ENST00000331782.8
ENST00000347004.2 |
JAG2
|
jagged canonical Notch ligand 2 |
| chr3_+_189789643 | 0.59 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chr3_-_185825029 | 0.59 |
ENST00000382199.7
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr14_+_93430927 | 0.56 |
ENST00000393151.6
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr3_-_185824966 | 0.54 |
ENST00000457616.6
ENST00000346192.7 |
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr20_-_22584547 | 0.53 |
ENST00000419308.7
|
FOXA2
|
forkhead box A2 |
| chr6_-_33317728 | 0.49 |
ENST00000431845.3
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr6_-_32190170 | 0.48 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2 |
| chr14_-_21526391 | 0.47 |
ENST00000611430.4
|
SALL2
|
spalt like transcription factor 2 |
| chr14_-_21526312 | 0.40 |
ENST00000537235.2
|
SALL2
|
spalt like transcription factor 2 |
| chr1_-_32964685 | 0.40 |
ENST00000373456.11
ENST00000356990.9 ENST00000235150.5 |
RNF19B
|
ring finger protein 19B |
| chr17_+_7439504 | 0.40 |
ENST00000575331.1
ENST00000293829.9 |
ENSG00000272884.1
FGF11
|
novel transcript fibroblast growth factor 11 |
| chr7_+_21428184 | 0.40 |
ENST00000649633.1
|
SP4
|
Sp4 transcription factor |
| chr17_+_57256727 | 0.37 |
ENST00000675656.1
|
MSI2
|
musashi RNA binding protein 2 |
| chr12_+_57755060 | 0.37 |
ENST00000266643.6
|
MARCHF9
|
membrane associated ring-CH-type finger 9 |
| chr19_+_45251363 | 0.35 |
ENST00000620044.4
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr12_-_54391270 | 0.34 |
ENST00000352268.10
ENST00000549962.5 ENST00000338010.9 ENST00000550774.5 |
ZNF385A
|
zinc finger protein 385A |
| chr2_-_60550900 | 0.34 |
ENST00000643222.1
ENST00000643459.1 ENST00000489516.7 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr16_+_81444799 | 0.34 |
ENST00000537098.8
|
CMIP
|
c-Maf inducing protein |
| chr19_+_45251249 | 0.34 |
ENST00000262891.9
ENST00000300843.8 |
MARK4
|
microtubule affinity regulating kinase 4 |
| chr12_-_53220377 | 0.34 |
ENST00000543726.1
|
RARG
|
retinoic acid receptor gamma |
| chr20_-_52191697 | 0.31 |
ENST00000361387.6
|
ZFP64
|
ZFP64 zinc finger protein |
| chr1_+_26529745 | 0.31 |
ENST00000374168.7
ENST00000374166.8 |
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr8_+_32548303 | 0.31 |
ENST00000650967.1
|
NRG1
|
neuregulin 1 |
| chrX_-_129654946 | 0.31 |
ENST00000429967.3
|
APLN
|
apelin |
| chr9_-_23821275 | 0.30 |
ENST00000380110.8
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr6_-_106974721 | 0.30 |
ENST00000606017.2
ENST00000620405.1 |
CD24
|
CD24 molecule |
| chr3_+_189789734 | 0.29 |
ENST00000437221.5
ENST00000392463.6 ENST00000392461.7 ENST00000449992.5 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr11_+_64234569 | 0.28 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr7_+_21428023 | 0.28 |
ENST00000432066.2
ENST00000222584.8 |
SP4
|
Sp4 transcription factor |
| chr17_-_41518878 | 0.28 |
ENST00000254043.8
|
KRT15
|
keratin 15 |
| chr11_+_68312542 | 0.27 |
ENST00000294304.12
|
LRP5
|
LDL receptor related protein 5 |
| chr8_-_22133356 | 0.27 |
ENST00000680789.1
|
HR
|
HR lysine demethylase and nuclear receptor corepressor |
| chr18_-_76495191 | 0.27 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr14_-_23183641 | 0.27 |
ENST00000469263.5
ENST00000525062.1 ENST00000316902.12 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 member 8 |
| chrX_+_153687918 | 0.27 |
ENST00000253122.10
|
SLC6A8
|
solute carrier family 6 member 8 |
| chr7_+_69598465 | 0.26 |
ENST00000342771.10
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr9_-_122228845 | 0.26 |
ENST00000394319.8
ENST00000340587.7 |
LHX6
|
LIM homeobox 6 |
| chr6_-_142945160 | 0.26 |
ENST00000367603.8
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr20_-_32483438 | 0.26 |
ENST00000359676.9
|
NOL4L
|
nucleolar protein 4 like |
| chr17_+_57256514 | 0.26 |
ENST00000284073.7
ENST00000674964.1 |
MSI2
|
musashi RNA binding protein 2 |
| chr16_+_29806078 | 0.25 |
ENST00000545521.5
|
MAZ
|
MYC associated zinc finger protein |
| chr19_+_46602050 | 0.25 |
ENST00000599839.5
ENST00000596362.1 |
CALM3
|
calmodulin 3 |
| chr6_-_142945028 | 0.25 |
ENST00000012134.7
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr6_+_149317530 | 0.25 |
ENST00000636456.1
|
TAB2
|
TGF-beta activated kinase 1 (MAP3K7) binding protein 2 |
| chr8_+_32548210 | 0.25 |
ENST00000523079.5
ENST00000650919.1 |
NRG1
|
neuregulin 1 |
| chr20_+_36573458 | 0.24 |
ENST00000373874.6
|
TGIF2
|
TGFB induced factor homeobox 2 |
| chr19_+_33796268 | 0.24 |
ENST00000587559.5
ENST00000588637.5 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr6_-_32192630 | 0.23 |
ENST00000375040.8
|
GPSM3
|
G protein signaling modulator 3 |
| chr6_-_34696839 | 0.23 |
ENST00000374026.7
|
ILRUN
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr6_-_34696733 | 0.23 |
ENST00000374023.8
|
ILRUN
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr20_+_36573589 | 0.22 |
ENST00000373872.9
ENST00000650844.1 |
TGIF2
|
TGFB induced factor homeobox 2 |
| chr3_-_171460368 | 0.21 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr6_+_149317695 | 0.21 |
ENST00000637181.2
|
TAB2
|
TGF-beta activated kinase 1 (MAP3K7) binding protein 2 |
| chr6_-_32192845 | 0.21 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3 |
| chr16_+_74999312 | 0.20 |
ENST00000566250.5
ENST00000567962.5 |
ZNRF1
|
zinc and ring finger 1 |
| chr13_-_40666600 | 0.20 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chrX_+_71095838 | 0.20 |
ENST00000374259.8
|
FOXO4
|
forkhead box O4 |
| chr19_-_43639788 | 0.20 |
ENST00000222374.3
|
CADM4
|
cell adhesion molecule 4 |
| chr12_+_57522801 | 0.20 |
ENST00000355673.8
ENST00000546632.1 ENST00000549623.1 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr12_-_57846686 | 0.20 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr19_-_18542971 | 0.19 |
ENST00000596558.6
|
FKBP8
|
FKBP prolyl isomerase 8 |
| chr17_+_59220446 | 0.19 |
ENST00000284116.9
ENST00000581140.5 ENST00000581276.5 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr19_-_18543556 | 0.19 |
ENST00000544835.7
ENST00000608443.6 ENST00000597960.7 |
FKBP8
|
FKBP prolyl isomerase 8 |
| chr19_-_18543483 | 0.19 |
ENST00000597547.1
ENST00000222308.8 |
FKBP8
|
FKBP prolyl isomerase 8 |
| chr12_-_54385727 | 0.19 |
ENST00000551109.5
ENST00000546970.5 |
ZNF385A
|
zinc finger protein 385A |
| chr19_-_48511793 | 0.19 |
ENST00000600059.6
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr10_-_101775974 | 0.18 |
ENST00000346714.7
ENST00000347978.2 |
FGF8
|
fibroblast growth factor 8 |
| chr8_+_122781621 | 0.18 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr6_-_42451261 | 0.18 |
ENST00000372917.8
ENST00000340840.6 ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr19_-_40716869 | 0.18 |
ENST00000677018.1
ENST00000324464.8 ENST00000594720.6 ENST00000677496.1 |
COQ8B
|
coenzyme Q8B |
| chr3_-_64687992 | 0.18 |
ENST00000498707.5
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr3_-_171460063 | 0.18 |
ENST00000284483.12
ENST00000475336.5 ENST00000357327.9 ENST00000460047.5 ENST00000488470.5 ENST00000470834.5 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr20_+_6767678 | 0.18 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2 |
| chr19_+_11346556 | 0.17 |
ENST00000587531.5
|
CCDC159
|
coiled-coil domain containing 159 |
| chr13_+_46553157 | 0.17 |
ENST00000311191.10
ENST00000389797.8 ENST00000389798.7 |
LRCH1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr17_-_7404039 | 0.17 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr1_-_242524687 | 0.17 |
ENST00000442594.6
ENST00000536534.7 |
PLD5
|
phospholipase D family member 5 |
| chr1_+_212285383 | 0.17 |
ENST00000261461.7
|
PPP2R5A
|
protein phosphatase 2 regulatory subunit B'alpha |
| chr1_-_18956669 | 0.17 |
ENST00000455833.7
|
IFFO2
|
intermediate filament family orphan 2 |
| chr17_-_76537699 | 0.16 |
ENST00000293230.10
|
CYGB
|
cytoglobin |
| chr3_-_133895453 | 0.16 |
ENST00000486858.5
ENST00000477759.5 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr1_-_40665654 | 0.16 |
ENST00000372684.8
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr15_-_42920638 | 0.16 |
ENST00000566931.1
ENST00000564431.5 ENST00000567274.5 ENST00000267890.11 |
TTBK2
|
tau tubulin kinase 2 |
| chr19_+_33796846 | 0.16 |
ENST00000590771.5
ENST00000589786.5 ENST00000284006.10 ENST00000683859.1 ENST00000588881.5 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr17_-_44199834 | 0.15 |
ENST00000587097.6
|
ATXN7L3
|
ataxin 7 like 3 |
| chr1_+_13584262 | 0.15 |
ENST00000376061.8
ENST00000513143.5 |
PDPN
|
podoplanin |
| chr20_-_32483507 | 0.15 |
ENST00000326071.8
|
NOL4L
|
nucleolar protein 4 like |
| chr1_+_65147830 | 0.15 |
ENST00000395334.6
|
AK4
|
adenylate kinase 4 |
| chr2_+_180981108 | 0.15 |
ENST00000602710.5
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3 |
| chr17_-_7217206 | 0.15 |
ENST00000447163.6
ENST00000647975.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr22_+_46150590 | 0.15 |
ENST00000262735.9
ENST00000420804.5 |
PPARA
|
peroxisome proliferator activated receptor alpha |
| chr14_+_95876385 | 0.15 |
ENST00000504119.1
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA |
| chrX_-_47650488 | 0.14 |
ENST00000247161.7
ENST00000376983.8 ENST00000343894.8 |
ELK1
|
ETS transcription factor ELK1 |
| chr1_-_6261053 | 0.14 |
ENST00000377893.3
|
GPR153
|
G protein-coupled receptor 153 |
| chr19_-_51646800 | 0.14 |
ENST00000599649.5
ENST00000429354.3 ENST00000360844.6 |
SIGLEC5
SIGLEC14
|
sialic acid binding Ig like lectin 5 sialic acid binding Ig like lectin 14 |
| chr7_+_107168961 | 0.14 |
ENST00000468410.5
ENST00000478930.5 ENST00000464009.1 ENST00000222574.9 |
HBP1
|
HMG-box transcription factor 1 |
| chr12_-_6689359 | 0.14 |
ENST00000683879.1
|
ZNF384
|
zinc finger protein 384 |
| chr18_-_26549402 | 0.14 |
ENST00000408011.7
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr7_-_105691637 | 0.14 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7 like 1 |
| chr12_+_7189845 | 0.14 |
ENST00000412720.6
ENST00000396637.7 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr3_-_142028597 | 0.14 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chr18_-_55588184 | 0.14 |
ENST00000354452.8
ENST00000565908.6 ENST00000635822.2 |
TCF4
|
transcription factor 4 |
| chr3_+_51983438 | 0.14 |
ENST00000476351.5
ENST00000476854.5 ENST00000494103.5 ENST00000404366.7 ENST00000635797.1 ENST00000636358.2 ENST00000469863.1 |
ACY1
|
aminoacylase 1 |
| chr1_+_95117324 | 0.14 |
ENST00000370203.9
ENST00000456991.5 |
TLCD4
|
TLC domain containing 4 |
| chr3_-_18424533 | 0.14 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr2_+_24049673 | 0.14 |
ENST00000380991.8
|
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr10_+_119207560 | 0.13 |
ENST00000392870.3
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr18_-_51197671 | 0.13 |
ENST00000406189.4
|
MEX3C
|
mex-3 RNA binding family member C |
| chr14_+_64704380 | 0.13 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr12_-_26125023 | 0.13 |
ENST00000242728.5
|
BHLHE41
|
basic helix-loop-helix family member e41 |
| chr16_-_4614859 | 0.13 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 |
UBALD1
|
UBA like domain containing 1 |
| chr1_-_37808168 | 0.13 |
ENST00000373044.3
|
YRDC
|
yrdC N6-threonylcarbamoyltransferase domain containing |
| chr7_+_151086466 | 0.13 |
ENST00000397238.7
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr6_+_27824084 | 0.13 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr7_+_20330893 | 0.13 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr16_+_29806519 | 0.13 |
ENST00000322945.11
ENST00000562337.5 ENST00000566906.6 ENST00000563402.1 ENST00000219782.10 |
MAZ
|
MYC associated zinc finger protein |
| chr2_+_219460719 | 0.13 |
ENST00000396688.5
|
SPEG
|
striated muscle enriched protein kinase |
| chr1_-_204152010 | 0.13 |
ENST00000367202.9
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_+_13583762 | 0.13 |
ENST00000376057.8
ENST00000621990.5 ENST00000510906.5 |
PDPN
|
podoplanin |
| chr12_-_53220229 | 0.13 |
ENST00000338561.9
|
RARG
|
retinoic acid receptor gamma |
| chr14_+_95876762 | 0.13 |
ENST00000503525.2
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA |
| chr1_-_243850070 | 0.13 |
ENST00000366539.6
ENST00000672578.1 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr4_+_145482761 | 0.13 |
ENST00000507367.1
ENST00000394092.6 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr3_+_194136138 | 0.13 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr9_+_33817126 | 0.13 |
ENST00000263228.4
|
UBE2R2
|
ubiquitin conjugating enzyme E2 R2 |
| chr17_+_56593685 | 0.12 |
ENST00000332822.6
|
NOG
|
noggin |
| chr8_+_123416735 | 0.12 |
ENST00000524254.5
|
NTAQ1
|
N-terminal glutamine amidase 1 |
| chr17_-_7251691 | 0.12 |
ENST00000574322.6
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chrX_+_77447387 | 0.12 |
ENST00000439435.3
|
FGF16
|
fibroblast growth factor 16 |
| chr15_-_42920798 | 0.12 |
ENST00000622375.4
ENST00000567840.5 |
TTBK2
|
tau tubulin kinase 2 |
| chr14_-_77141777 | 0.12 |
ENST00000319374.4
|
ZDHHC22
|
zinc finger DHHC-type palmitoyltransferase 22 |
| chr18_+_58862904 | 0.12 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532 |
| chr12_-_6689244 | 0.12 |
ENST00000361959.7
ENST00000436774.6 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr1_-_204151884 | 0.12 |
ENST00000367201.7
|
ETNK2
|
ethanolamine kinase 2 |
| chr6_-_42451910 | 0.12 |
ENST00000372922.8
ENST00000541110.5 |
TRERF1
|
transcriptional regulating factor 1 |
| chr22_-_39244969 | 0.12 |
ENST00000331163.11
|
PDGFB
|
platelet derived growth factor subunit B |
| chr10_-_118754956 | 0.12 |
ENST00000369151.8
|
CACUL1
|
CDK2 associated cullin domain 1 |
| chr2_+_24049705 | 0.12 |
ENST00000380986.9
ENST00000452109.1 |
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr16_+_67029359 | 0.12 |
ENST00000565389.1
|
CBFB
|
core-binding factor subunit beta |
| chr3_+_50155305 | 0.12 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr1_+_167936818 | 0.12 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr10_-_84241538 | 0.12 |
ENST00000372105.4
|
LRIT1
|
leucine rich repeat, Ig-like and transmembrane domains 1 |
| chr19_-_45973863 | 0.12 |
ENST00000263257.6
|
NOVA2
|
NOVA alternative splicing regulator 2 |
| chr1_+_161159450 | 0.12 |
ENST00000492950.5
ENST00000289865.12 ENST00000368002.8 ENST00000479344.1 ENST00000368001.1 |
USP21
|
ubiquitin specific peptidase 21 |
| chr17_+_35809474 | 0.12 |
ENST00000604879.5
ENST00000603427.5 ENST00000603777.5 ENST00000605844.6 ENST00000604841.5 |
TAF15
|
TATA-box binding protein associated factor 15 |
| chr10_+_97572771 | 0.12 |
ENST00000370655.6
ENST00000455090.1 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr8_+_123416718 | 0.12 |
ENST00000523984.5
|
NTAQ1
|
N-terminal glutamine amidase 1 |
| chr12_+_51239278 | 0.11 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr17_-_44199206 | 0.11 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7 like 3 |
| chr2_+_28751802 | 0.11 |
ENST00000296122.10
ENST00000395366.3 |
PPP1CB
|
protein phosphatase 1 catalytic subunit beta |
| chr10_-_101776104 | 0.11 |
ENST00000320185.7
ENST00000344255.8 |
FGF8
|
fibroblast growth factor 8 |
| chr3_+_50155024 | 0.11 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr15_-_88256158 | 0.11 |
ENST00000317501.7
ENST00000629765.2 ENST00000557856.5 ENST00000558676.5 |
NTRK3
|
neurotrophic receptor tyrosine kinase 3 |
| chr17_-_29589606 | 0.11 |
ENST00000225394.8
|
GIT1
|
GIT ArfGAP 1 |
| chr16_+_11668414 | 0.11 |
ENST00000329565.6
|
SNN
|
stannin |
| chrX_+_136148440 | 0.11 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
| chr19_-_14090695 | 0.11 |
ENST00000533683.7
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr6_-_170291053 | 0.11 |
ENST00000366756.4
|
DLL1
|
delta like canonical Notch ligand 1 |
| chr11_-_123654581 | 0.11 |
ENST00000392770.6
ENST00000530277.5 ENST00000299333.8 |
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr5_-_95961830 | 0.11 |
ENST00000513343.1
ENST00000237853.9 |
ELL2
|
elongation factor for RNA polymerase II 2 |
| chr16_+_3024000 | 0.11 |
ENST00000326266.13
ENST00000574549.5 ENST00000575576.5 ENST00000253952.9 |
THOC6
|
THO complex 6 |
| chr16_-_3024230 | 0.11 |
ENST00000572355.5
ENST00000574980.5 ENST00000354679.3 ENST00000573842.1 |
HCFC1R1
|
host cell factor C1 regulator 1 |
| chrX_-_48971829 | 0.11 |
ENST00000218176.4
|
KCND1
|
potassium voltage-gated channel subfamily D member 1 |
| chr6_+_18155399 | 0.11 |
ENST00000650836.2
ENST00000449850.2 ENST00000297792.9 |
KDM1B
|
lysine demethylase 1B |
| chr2_-_148021490 | 0.11 |
ENST00000416719.5
ENST00000264169.6 |
ORC4
|
origin recognition complex subunit 4 |
| chr18_+_3449817 | 0.11 |
ENST00000407501.6
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr17_-_65056659 | 0.10 |
ENST00000439174.7
|
GNA13
|
G protein subunit alpha 13 |
| chr14_+_93430853 | 0.10 |
ENST00000553484.5
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr8_+_123416766 | 0.10 |
ENST00000287387.7
ENST00000650311.1 ENST00000523356.1 |
NTAQ1
|
N-terminal glutamine amidase 1 |
| chr17_+_82032182 | 0.10 |
ENST00000584341.1
|
RAC3
|
Rac family small GTPase 3 |
| chr12_+_7189675 | 0.10 |
ENST00000675855.1
ENST00000434354.6 ENST00000544456.5 ENST00000545574.5 ENST00000420616.6 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr18_+_79964629 | 0.10 |
ENST00000451882.3
|
HSBP1L1
|
heat shock factor binding protein 1 like 1 |
| chr15_-_45187955 | 0.10 |
ENST00000560471.5
ENST00000560540.5 |
SHF
|
Src homology 2 domain containing F |
| chr7_-_151814636 | 0.10 |
ENST00000652047.1
ENST00000650858.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr17_+_82032061 | 0.10 |
ENST00000580965.5
|
RAC3
|
Rac family small GTPase 3 |
| chr14_-_22982544 | 0.10 |
ENST00000262713.7
|
AJUBA
|
ajuba LIM protein |
| chr17_-_76537630 | 0.10 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr12_-_57520480 | 0.10 |
ENST00000642841.1
ENST00000547303.5 ENST00000552740.5 ENST00000547526.1 ENST00000346473.8 ENST00000551116.5 |
ENSG00000285133.1
DDIT3
|
novel protein DNA damage inducible transcript 3 |
| chr9_-_37465402 | 0.10 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr19_-_17245889 | 0.10 |
ENST00000291442.4
|
NR2F6
|
nuclear receptor subfamily 2 group F member 6 |
| chr8_+_26383043 | 0.10 |
ENST00000380629.7
|
BNIP3L
|
BCL2 interacting protein 3 like |
| chr6_+_42782020 | 0.10 |
ENST00000314073.9
|
BICRAL
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr16_+_67029093 | 0.10 |
ENST00000561924.6
|
CBFB
|
core-binding factor subunit beta |
| chr7_+_26201705 | 0.10 |
ENST00000396386.7
ENST00000456948.5 ENST00000409747.5 |
CBX3
|
chromobox 3 |
| chr12_-_6689450 | 0.10 |
ENST00000355772.8
ENST00000417772.7 ENST00000319770.7 ENST00000396801.7 |
ZNF384
|
zinc finger protein 384 |
| chr14_-_21094488 | 0.10 |
ENST00000555270.5
|
ZNF219
|
zinc finger protein 219 |
| chr1_-_156500723 | 0.10 |
ENST00000368240.6
|
MEF2D
|
myocyte enhancer factor 2D |
| chr1_-_156500763 | 0.10 |
ENST00000348159.9
ENST00000489057.1 |
MEF2D
|
myocyte enhancer factor 2D |
| chr19_-_45973986 | 0.10 |
ENST00000676183.1
|
NOVA2
|
NOVA alternative splicing regulator 2 |
| chr17_+_45161070 | 0.10 |
ENST00000593138.6
ENST00000586681.6 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chr6_+_33428223 | 0.09 |
ENST00000682587.1
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr15_-_45187947 | 0.09 |
ENST00000560734.5
|
SHF
|
Src homology 2 domain containing F |
| chr19_-_41397256 | 0.09 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.9 |
EXOSC5
|
exosome component 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF740_ZNF219
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 0.5 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.5 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 | 0.5 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.5 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.3 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 | 0.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.3 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.2 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.3 | GO:0032599 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.1 | 0.7 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.2 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.2 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.2 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.3 | GO:2000489 | regulation of hepatic stellate cell activation(GO:2000489) |
| 0.0 | 0.7 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.3 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.5 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.1 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.3 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 5.2 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:2000798 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.0 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.0 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.5 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.0 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.8 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 5.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |