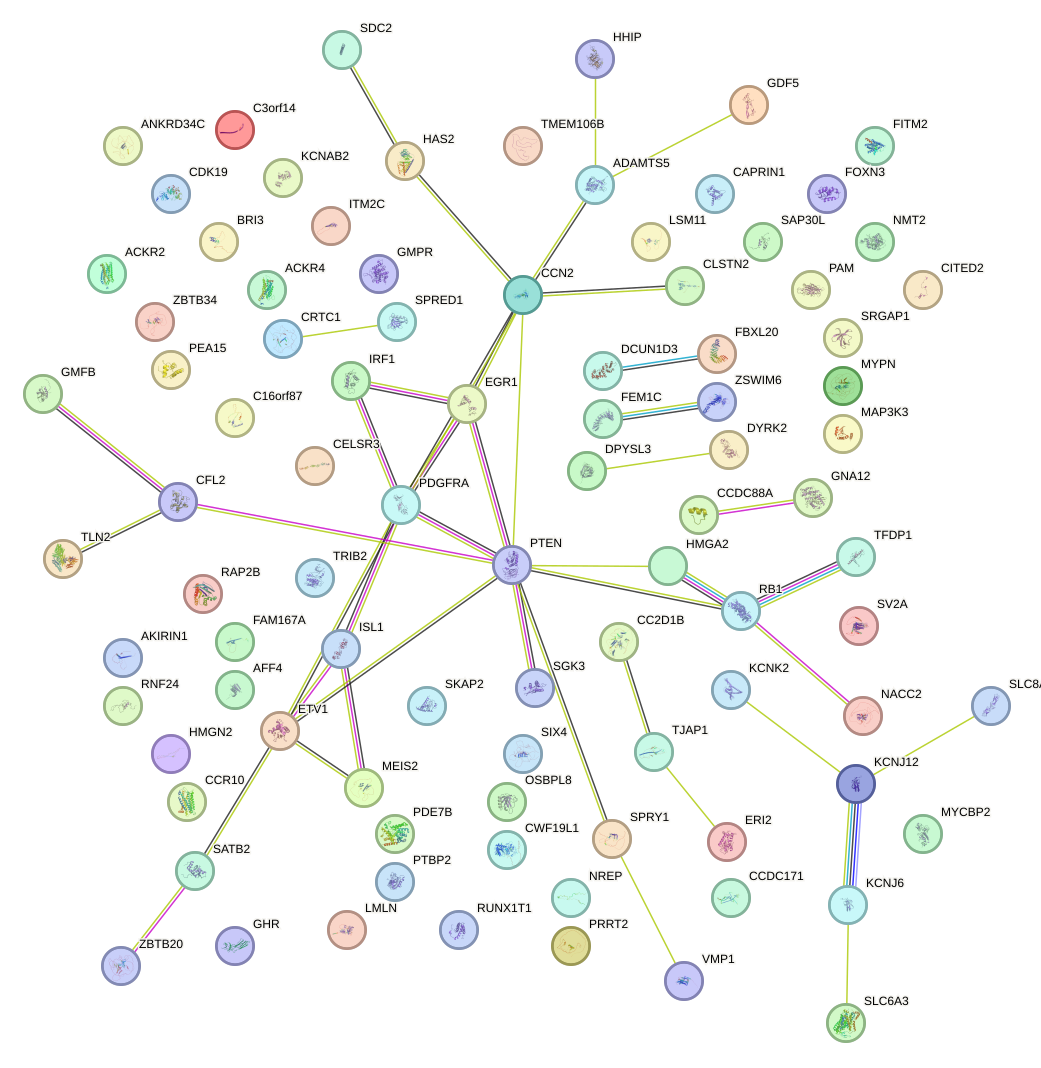
|
chr5_-_111757704
Show fit
|
0.68 |
ENST00000379671.7
|
NREP
|
neuronal regeneration related protein
|
|
chr4_+_144646145
Show fit
|
0.47 |
ENST00000296575.8
ENST00000434550.2
|
HHIP
|
hedgehog interacting protein
|
|
chr4_+_123399488
Show fit
|
0.44 |
ENST00000394339.2
|
SPRY1
|
sprouty RTK signaling antagonist 1
|
|
chr5_-_147453888
Show fit
|
0.42 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3
|
|
chr1_+_215082731
Show fit
|
0.37 |
ENST00000444842.7
|
KCNK2
|
potassium two pore domain channel subfamily K member 2
|
|
chr3_-_115071333
Show fit
|
0.37 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_+_96493803
Show fit
|
0.37 |
ENST00000518385.5
ENST00000302190.9
|
SDC2
|
syndecan 2
|
|
chr11_-_70661762
Show fit
|
0.36 |
ENST00000357171.7
ENST00000412252.5
ENST00000449833.6
ENST00000338508.8
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr6_+_108559742
Show fit
|
0.29 |
ENST00000343882.10
|
FOXO3
|
forkhead box O3
|
|
chr9_-_136050502
Show fit
|
0.28 |
ENST00000371753.5
|
NACC2
|
NACC family member 2
|
|
chr14_-_29927801
Show fit
|
0.25 |
ENST00000331968.11
|
PRKD1
|
protein kinase D1
|
|
chr14_-_89619118
Show fit
|
0.25 |
ENST00000345097.8
ENST00000555855.5
ENST00000555353.5
|
FOXN3
|
forkhead box N3
|
|
chr10_+_68109433
Show fit
|
0.25 |
ENST00000613327.4
ENST00000358913.10
ENST00000373675.3
|
MYPN
|
myopalladin
|
|
chr5_+_154445979
Show fit
|
0.22 |
ENST00000297109.11
|
SAP30L
|
SAP30 like
|
|
chr8_-_92103217
Show fit
|
0.21 |
ENST00000615601.4
ENST00000523629.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1
|
|
chr6_-_131951364
Show fit
|
0.20 |
ENST00000367976.4
|
CCN2
|
cellular communication network factor 2
|
|
chr6_-_139374605
Show fit
|
0.20 |
ENST00000618718.1
ENST00000367651.4
|
CITED2
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 2
|
|
chr3_-_48662877
Show fit
|
0.18 |
ENST00000164024.5
|
CELSR3
|
cadherin EGF LAG seven-pass G-type receptor 3
|
|
chr21_-_37916440
Show fit
|
0.18 |
ENST00000609713.2
|
KCNJ6
|
potassium inwardly rectifying channel subfamily J member 6
|
|
chr5_-_115544734
Show fit
|
0.18 |
ENST00000274457.5
|
FEM1C
|
fem-1 homolog C
|
|
chr6_+_135851681
Show fit
|
0.15 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B
|
|
chr5_+_157743703
Show fit
|
0.15 |
ENST00000286307.6
|
LSM11
|
LSM11, U7 small nuclear RNA associated
|
|
chr17_+_63622406
Show fit
|
0.14 |
ENST00000579585.5
ENST00000361733.8
ENST00000584573.5
ENST00000361357.7
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr6_+_16238554
Show fit
|
0.13 |
ENST00000259727.5
|
GMPR
|
guanosine monophosphate reductase
|
|
chr1_+_96721762
Show fit
|
0.13 |
ENST00000675735.1
ENST00000609116.5
ENST00000674951.1
ENST00000426398.3
ENST00000370197.5
ENST00000370198.5
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr6_-_110815408
Show fit
|
0.13 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19
|
|
chr2_-_199457931
Show fit
|
0.13 |
ENST00000417098.6
|
SATB2
|
SATB homeobox 2
|
|
chr17_+_21376321
Show fit
|
0.12 |
ENST00000583088.6
|
KCNJ12
|
potassium inwardly rectifying channel subfamily J member 12
|
|
chr8_-_121641424
Show fit
|
0.12 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2
|
|
chr16_+_29812230
Show fit
|
0.12 |
ENST00000300797.7
ENST00000637403.1
ENST00000572820.2
ENST00000637064.1
ENST00000636246.1
|
PRRT2
|
proline rich transmembrane protein 2
|
|
chr15_+_38252792
Show fit
|
0.11 |
ENST00000299084.9
|
SPRED1
|
sprouty related EVH1 domain containing 1
|
|
chr14_-_34713788
Show fit
|
0.11 |
ENST00000341223.8
|
CFL2
|
cofilin 2
|
|
chr20_-_35438218
Show fit
|
0.11 |
ENST00000374369.8
|
GDF5
|
growth differentiation factor 5
|
|
chr15_+_62561361
Show fit
|
0.10 |
ENST00000561311.5
|
TLN2
|
talin 2
|
|
chr3_+_197960200
Show fit
|
0.10 |
ENST00000482695.5
ENST00000330198.8
ENST00000419117.5
ENST00000420910.6
ENST00000332636.5
|
LMLN
|
leishmanolysin like peptidase
|
|
chr20_-_4015389
Show fit
|
0.10 |
ENST00000336095.10
|
RNF24
|
ring finger protein 24
|
|
chr21_-_26967057
Show fit
|
0.10 |
ENST00000284987.6
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif 5
|
|
chr10_-_15168667
Show fit
|
0.09 |
ENST00000378165.9
|
NMT2
|
N-myristoyltransferase 2
|
|
chr5_+_61332236
Show fit
|
0.09 |
ENST00000252744.6
|
ZSWIM6
|
zinc finger SWIM-type containing 6
|
|
chr3_-_64445396
Show fit
|
0.09 |
ENST00000295902.11
|
PRICKLE2
|
prickle planar cell polarity protein 2
|
|
chr2_-_55419565
Show fit
|
0.09 |
ENST00000647341.1
ENST00000647401.1
ENST00000336838.10
ENST00000621814.4
ENST00000644033.1
ENST00000645477.1
ENST00000647517.1
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr17_+_59707636
Show fit
|
0.08 |
ENST00000262291.9
ENST00000587945.1
ENST00000589823.6
ENST00000592106.5
ENST00000591315.5
|
VMP1
|
vacuole membrane protein 1
|
|
chr7_+_98281669
Show fit
|
0.08 |
ENST00000297290.4
|
BRI3
|
brain protein I3
|
|
chr15_+_79282709
Show fit
|
0.08 |
ENST00000421388.4
|
ANKRD34C
|
ankyrin repeat domain 34C
|
|
chr12_+_63844663
Show fit
|
0.08 |
ENST00000355086.8
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr4_+_54229261
Show fit
|
0.08 |
ENST00000508170.5
ENST00000512143.1
ENST00000257290.10
|
PDGFRA
|
platelet derived growth factor receptor alpha
|
|
chr1_-_149917826
Show fit
|
0.08 |
ENST00000369145.1
ENST00000369146.8
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr8_-_11466740
Show fit
|
0.08 |
ENST00000284486.9
|
FAM167A
|
family with sequence similarity 167 member A
|
|
chr5_+_42423433
Show fit
|
0.07 |
ENST00000230882.9
|
GHR
|
growth hormone receptor
|
|
chr12_+_65824475
Show fit
|
0.07 |
ENST00000403681.7
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr2_+_12716893
Show fit
|
0.07 |
ENST00000381465.2
ENST00000155926.9
|
TRIB2
|
tribbles pseudokinase 2
|
|
chr10_+_87863595
Show fit
|
0.07 |
ENST00000371953.8
|
PTEN
|
phosphatase and tensin homolog
|
|
chr2_-_98936155
Show fit
|
0.07 |
ENST00000428096.5
ENST00000397899.7
ENST00000420294.1
|
CRACDL
|
CRACD like
|
|
chr3_+_139935176
Show fit
|
0.07 |
ENST00000458420.7
|
CLSTN2
|
calsyntenin 2
|
|
chr16_-_46831134
Show fit
|
0.07 |
ENST00000394806.6
ENST00000285697.9
|
C16orf87
|
chromosome 16 open reading frame 87
|
|
chr9_+_126860625
Show fit
|
0.07 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr13_+_113584683
Show fit
|
0.06 |
ENST00000375370.10
|
TFDP1
|
transcription factor Dp-1
|
|
chr11_+_34051722
Show fit
|
0.06 |
ENST00000341394.9
ENST00000389645.7
|
CAPRIN1
|
cell cycle associated protein 1
|
|
chr16_-_20900319
Show fit
|
0.06 |
ENST00000564349.5
ENST00000324344.9
|
ERI2
DCUN1D3
|
ERI1 exoribonuclease family member 2
defective in cullin neddylation 1 domain containing 3
|
|
chr20_-_44311142
Show fit
|
0.06 |
ENST00000396825.4
|
FITM2
|
fat storage inducing transmembrane protein 2
|
|
chr5_+_102755269
Show fit
|
0.05 |
ENST00000304400.12
ENST00000455264.7
ENST00000684529.1
ENST00000438793.8
ENST00000682882.1
ENST00000682972.1
ENST00000348126.7
ENST00000512073.1
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr12_-_76559504
Show fit
|
0.05 |
ENST00000547544.5
ENST00000393249.6
|
OSBPL8
|
oxysterol binding protein like 8
|
|
chr9_+_15553002
Show fit
|
0.05 |
ENST00000380701.8
|
CCDC171
|
coiled-coil domain containing 171
|
|
chr1_-_52366124
Show fit
|
0.05 |
ENST00000371586.6
ENST00000284376.8
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr14_-_60724300
Show fit
|
0.05 |
ENST00000556952.3
ENST00000216513.5
|
SIX4
|
SIX homeobox 4
|
|
chr7_-_112206380
Show fit
|
0.05 |
ENST00000437633.6
ENST00000428084.6
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr3_+_62319037
Show fit
|
0.05 |
ENST00000494481.5
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr11_-_12009082
Show fit
|
0.05 |
ENST00000396505.7
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3
|
|
chr7_-_2844294
Show fit
|
0.05 |
ENST00000275364.8
|
GNA12
|
G protein subunit alpha 12
|
|
chr16_+_69565958
Show fit
|
0.05 |
ENST00000349945.7
ENST00000354436.6
|
NFAT5
|
nuclear factor of activated T cells 5
|
|
chr5_-_1445424
Show fit
|
0.04 |
ENST00000270349.12
|
SLC6A3
|
solute carrier family 6 member 3
|
|
chr7_-_13989658
Show fit
|
0.04 |
ENST00000430479.6
ENST00000433547.1
ENST00000405192.6
|
ETV1
|
ETS variant transcription factor 1
|
|
chr19_+_18683656
Show fit
|
0.04 |
ENST00000338797.10
ENST00000321949.13
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr14_-_54489003
Show fit
|
0.04 |
ENST00000554908.5
ENST00000616146.4
|
GMFB
|
glia maturation factor beta
|
|
chr7_-_26864573
Show fit
|
0.04 |
ENST00000345317.7
|
SKAP2
|
src kinase associated phosphoprotein 2
|
|
chr13_+_48303709
Show fit
|
0.04 |
ENST00000646097.1
ENST00000650461.1
ENST00000267163.6
|
RB1
|
RB transcriptional corepressor 1
|
|
chr3_+_153162196
Show fit
|
0.04 |
ENST00000323534.5
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr10_-_100267598
Show fit
|
0.04 |
ENST00000354105.10
ENST00000473842.1
|
CWF19L1
|
CWF19 like cell cycle control factor 1
|
|
chr17_-_39401593
Show fit
|
0.04 |
ENST00000394294.7
ENST00000264658.11
ENST00000583610.5
ENST00000647139.1
|
FBXL20
|
F-box and leucine rich repeat protein 20
|
|
chr1_+_38991239
Show fit
|
0.04 |
ENST00000432648.8
ENST00000446189.6
ENST00000372984.8
|
AKIRIN1
|
akirin 1
|
|
chr7_+_12211259
Show fit
|
0.04 |
ENST00000396668.8
ENST00000444443.5
ENST00000396667.7
|
TMEM106B
|
transmembrane protein 106B
|
|
chr1_+_160205374
Show fit
|
0.04 |
ENST00000368077.5
ENST00000360472.9
|
PEA15
|
proliferation and apoptosis adaptor protein 15
|
|
chr5_+_138465472
Show fit
|
0.04 |
ENST00000239938.5
|
EGR1
|
early growth response 1
|
|
chr1_-_100894775
Show fit
|
0.04 |
ENST00000416479.1
ENST00000370113.7
|
EXTL2
|
exostosin like glycosyltransferase 2
|
|
chr8_+_66667572
Show fit
|
0.04 |
ENST00000520044.5
ENST00000519289.1
ENST00000521113.1
ENST00000661636.1
ENST00000521889.5
|
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough
chromosome 8 open reading frame 44
|
|
chr17_-_42681840
Show fit
|
0.03 |
ENST00000332438.4
|
CCR10
|
C-C motif chemokine receptor 10
|
|
chr1_+_26472405
Show fit
|
0.03 |
ENST00000361427.6
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr2_-_40452046
Show fit
|
0.03 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1
|
|
chr12_+_67648737
Show fit
|
0.03 |
ENST00000344096.4
ENST00000393555.3
|
DYRK2
|
dual specificity tyrosine phosphorylation regulated kinase 2
|
|
chr6_+_43477523
Show fit
|
0.03 |
ENST00000372444.6
ENST00000372445.9
ENST00000436109.6
ENST00000372454.6
ENST00000442878.6
ENST00000259751.5
ENST00000372452.5
ENST00000372449.5
|
TJAP1
|
tight junction associated protein 1
|
|
chr5_+_51383394
Show fit
|
0.03 |
ENST00000230658.12
|
ISL1
|
ISL LIM homeobox 1
|
|
chr5_-_132490750
Show fit
|
0.03 |
ENST00000437654.6
ENST00000245414.9
ENST00000680139.1
ENST00000680352.1
ENST00000679440.1
ENST00000680903.1
|
IRF1
|
interferon regulatory factor 1
|
|
chr5_-_132963621
Show fit
|
0.03 |
ENST00000265343.10
|
AFF4
|
AF4/FMR2 family member 4
|
|
chr15_-_37098281
Show fit
|
0.03 |
ENST00000559085.5
ENST00000397624.7
|
MEIS2
|
Meis homeobox 2
|
|
chr20_-_5001474
Show fit
|
0.03 |
ENST00000338244.6
|
SLC23A2
|
solute carrier family 23 member 2
|
|
chr15_+_44427591
Show fit
|
0.03 |
ENST00000558791.5
ENST00000260327.9
|
CTDSPL2
|
CTD small phosphatase like 2
|
|
chr1_-_16352420
Show fit
|
0.03 |
ENST00000375592.8
|
FBXO42
|
F-box protein 42
|
|
chrX_-_154097668
Show fit
|
0.03 |
ENST00000407218.5
ENST00000303391.11
ENST00000453960.7
|
MECP2
|
methyl-CpG binding protein 2
|
|
chr5_+_123089223
Show fit
|
0.03 |
ENST00000407847.5
|
PRDM6
|
PR/SET domain 6
|
|
chr10_-_32347109
Show fit
|
0.03 |
ENST00000469059.2
ENST00000319778.11
|
EPC1
|
enhancer of polycomb homolog 1
|
|
chr3_+_186783567
Show fit
|
0.03 |
ENST00000323963.10
ENST00000440191.6
|
EIF4A2
|
eukaryotic translation initiation factor 4A2
|
|
chr18_-_21111778
Show fit
|
0.02 |
ENST00000399799.3
|
ROCK1
|
Rho associated coiled-coil containing protein kinase 1
|
|
chr18_-_26090584
Show fit
|
0.02 |
ENST00000415083.7
|
SS18
|
SS18 subunit of BAF chromatin remodeling complex
|
|
chr3_+_43690880
Show fit
|
0.02 |
ENST00000458276.7
|
ABHD5
|
abhydrolase domain containing 5, lysophosphatidic acid acyltransferase
|
|
chr4_+_143184910
Show fit
|
0.02 |
ENST00000510377.5
ENST00000307017.9
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr17_-_1179940
Show fit
|
0.02 |
ENST00000302538.10
|
ABR
|
ABR activator of RhoGEF and GTPase
|
|
chr18_+_905103
Show fit
|
0.02 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1
|
|
chr1_-_235328147
Show fit
|
0.02 |
ENST00000264183.9
ENST00000418304.1
ENST00000349213.7
|
ARID4B
|
AT-rich interaction domain 4B
|
|
chr3_+_49554436
Show fit
|
0.02 |
ENST00000296452.5
|
BSN
|
bassoon presynaptic cytomatrix protein
|
|
chr1_+_202122910
Show fit
|
0.02 |
ENST00000682545.1
ENST00000367282.6
ENST00000682887.1
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chr12_-_117190456
Show fit
|
0.02 |
ENST00000330622.9
ENST00000622495.5
ENST00000427718.6
|
FBXO21
|
F-box protein 21
|
|
chr6_+_163414637
Show fit
|
0.02 |
ENST00000453779.6
ENST00000275262.11
ENST00000392127.6
|
QKI
|
QKI, KH domain containing RNA binding
|
|
chr10_-_13348270
Show fit
|
0.02 |
ENST00000378614.8
ENST00000327347.10
ENST00000545675.5
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr1_-_244864560
Show fit
|
0.02 |
ENST00000444376.7
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr17_+_28042660
Show fit
|
0.02 |
ENST00000407008.8
|
NLK
|
nemo like kinase
|
|
chr16_+_58463663
Show fit
|
0.02 |
ENST00000258187.9
|
NDRG4
|
NDRG family member 4
|
|
chr16_-_79600698
Show fit
|
0.02 |
ENST00000393350.1
|
MAF
|
MAF bZIP transcription factor
|
|
chr17_-_16215520
Show fit
|
0.02 |
ENST00000582357.5
ENST00000436828.5
ENST00000268712.8
ENST00000411510.5
ENST00000395857.7
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr10_+_67884646
Show fit
|
0.02 |
ENST00000212015.11
|
SIRT1
|
sirtuin 1
|
|
chr2_+_47783082
Show fit
|
0.02 |
ENST00000614496.4
ENST00000622629.4
ENST00000234420.11
ENST00000616033.4
ENST00000673637.1
|
MSH6
|
mutS homolog 6
|
|
chr5_+_136132772
Show fit
|
0.02 |
ENST00000545279.6
ENST00000507118.5
ENST00000511116.5
ENST00000545620.5
ENST00000509297.6
|
SMAD5
|
SMAD family member 5
|
|
chr2_+_176099787
Show fit
|
0.02 |
ENST00000406506.4
ENST00000404162.2
|
HOXD12
|
homeobox D12
|
|
chr10_+_112447198
Show fit
|
0.02 |
ENST00000393077.3
ENST00000432306.5
|
VTI1A
|
vesicle transport through interaction with t-SNAREs 1A
|
|
chr20_+_33490073
Show fit
|
0.02 |
ENST00000342704.11
ENST00000375279.6
|
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2
|
|
chr16_-_66751591
Show fit
|
0.02 |
ENST00000440564.6
ENST00000443351.6
ENST00000566150.5
ENST00000258198.7
|
DYNC1LI2
|
dynein cytoplasmic 1 light intermediate chain 2
|
|
chr6_+_133889105
Show fit
|
0.02 |
ENST00000367882.5
|
TCF21
|
transcription factor 21
|
|
chr2_+_156436423
Show fit
|
0.02 |
ENST00000540309.5
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2
|
|
chr20_+_58651228
Show fit
|
0.02 |
ENST00000361830.7
ENST00000458280.5
ENST00000355957.9
ENST00000312283.12
ENST00000412911.5
ENST00000359617.8
ENST00000371141.8
|
STX16
|
syntaxin 16
|
|
chr10_+_91923762
Show fit
|
0.02 |
ENST00000265990.11
|
BTAF1
|
B-TFIID TATA-box binding protein associated factor 1
|
|
chr1_+_61082553
Show fit
|
0.02 |
ENST00000403491.8
ENST00000371187.7
|
NFIA
|
nuclear factor I A
|
|
chr4_+_7043315
Show fit
|
0.02 |
ENST00000310074.8
ENST00000512388.1
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr7_+_114922346
Show fit
|
0.02 |
ENST00000393486.5
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr5_+_139342442
Show fit
|
0.02 |
ENST00000394795.6
ENST00000510080.1
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr22_+_20080211
Show fit
|
0.02 |
ENST00000383024.6
ENST00000351989.8
|
DGCR8
|
DGCR8 microprocessor complex subunit
|
|
chr15_+_72474305
Show fit
|
0.02 |
ENST00000379887.9
|
ARIH1
|
ariadne RBR E3 ubiquitin protein ligase 1
|
|
chr12_+_51238724
Show fit
|
0.02 |
ENST00000449723.7
ENST00000549555.5
ENST00000439799.6
ENST00000412716.8
ENST00000425012.6
|
DAZAP2
|
DAZ associated protein 2
|
|
chr11_+_18322541
Show fit
|
0.02 |
ENST00000534641.5
ENST00000265963.9
ENST00000525831.5
|
GTF2H1
|
general transcription factor IIH subunit 1
|
|
chr9_-_16870662
Show fit
|
0.02 |
ENST00000380672.9
|
BNC2
|
basonuclin 2
|
|
chr20_-_51542658
Show fit
|
0.01 |
ENST00000396009.7
ENST00000371564.8
ENST00000610033.5
|
NFATC2
|
nuclear factor of activated T cells 2
|
|
chr14_-_31207758
Show fit
|
0.01 |
ENST00000399332.6
ENST00000553700.5
|
HECTD1
|
HECT domain E3 ubiquitin protein ligase 1
|
|
chr9_-_14314067
Show fit
|
0.01 |
ENST00000397575.7
|
NFIB
|
nuclear factor I B
|
|
chr6_+_42782020
Show fit
|
0.01 |
ENST00000314073.9
|
BICRAL
|
BRD4 interacting chromatin remodeling complex associated protein like
|
|
chr3_+_37452121
Show fit
|
0.01 |
ENST00000264741.10
|
ITGA9
|
integrin subunit alpha 9
|
|
chr4_+_51843063
Show fit
|
0.01 |
ENST00000381441.7
ENST00000334635.10
|
DCUN1D4
|
defective in cullin neddylation 1 domain containing 4
|
|
chr17_+_32350132
Show fit
|
0.01 |
ENST00000321233.10
ENST00000394673.6
ENST00000394670.9
ENST00000579634.5
ENST00000580759.5
ENST00000342555.10
ENST00000577908.5
ENST00000394679.9
ENST00000582165.1
|
ZNF207
|
zinc finger protein 207
|
|
chr13_-_49936298
Show fit
|
0.01 |
ENST00000613924.1
ENST00000361840.8
|
SPRYD7
|
SPRY domain containing 7
|
|
chr4_-_70839873
Show fit
|
0.01 |
ENST00000545193.5
ENST00000254799.11
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr9_-_71768386
Show fit
|
0.01 |
ENST00000377066.9
ENST00000377044.9
|
CEMIP2
|
cell migration inducing hyaluronidase 2
|
|
chr12_+_22625075
Show fit
|
0.01 |
ENST00000671733.1
ENST00000335148.8
ENST00000672951.1
ENST00000266517.9
|
ETNK1
|
ethanolamine kinase 1
|
|
chr3_+_179148341
Show fit
|
0.01 |
ENST00000263967.4
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
|
|
chr6_+_87155537
Show fit
|
0.01 |
ENST00000369577.8
ENST00000518845.1
ENST00000339907.8
ENST00000496806.2
|
ZNF292
|
zinc finger protein 292
|
|
chr8_-_118621901
Show fit
|
0.01 |
ENST00000409003.5
ENST00000524796.6
ENST00000314727.9
|
SAMD12
|
sterile alpha motif domain containing 12
|
|
chr7_+_28412511
Show fit
|
0.01 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_+_36678699
Show fit
|
0.01 |
ENST00000405375.5
ENST00000244741.10
ENST00000373711.3
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A
|
|
chr17_+_45894515
Show fit
|
0.01 |
ENST00000680674.1
ENST00000535772.6
ENST00000351559.10
ENST00000262410.10
ENST00000344290.10
|
MAPT
|
microtubule associated protein tau
|
|
chr6_+_13615322
Show fit
|
0.01 |
ENST00000451315.7
|
NOL7
|
nucleolar protein 7
|
|
chr14_-_58427509
Show fit
|
0.01 |
ENST00000395159.7
|
TIMM9
|
translocase of inner mitochondrial membrane 9
|
|
chr3_-_122514876
Show fit
|
0.01 |
ENST00000493510.1
ENST00000476916.5
ENST00000344337.11
ENST00000465882.5
|
KPNA1
|
karyopherin subunit alpha 1
|
|
chr3_-_133895867
Show fit
|
0.01 |
ENST00000285208.9
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr1_-_23344314
Show fit
|
0.01 |
ENST00000374612.5
ENST00000675048.1
ENST00000478691.5
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr2_+_165239388
Show fit
|
0.01 |
ENST00000424833.5
ENST00000375437.7
ENST00000631182.3
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2
|
|
chr2_-_25252251
Show fit
|
0.01 |
ENST00000380746.8
ENST00000402667.1
|
DNMT3A
|
DNA methyltransferase 3 alpha
|
|
chr16_-_62036399
Show fit
|
0.01 |
ENST00000584337.5
|
CDH8
|
cadherin 8
|
|
chr15_-_68820861
Show fit
|
0.01 |
ENST00000560303.1
ENST00000465139.6
|
ANP32A
|
acidic nuclear phosphoprotein 32 family member A
|
|
chr14_-_63543328
Show fit
|
0.01 |
ENST00000337537.8
|
PPP2R5E
|
protein phosphatase 2 regulatory subunit B'epsilon
|
|
chr7_+_128739292
Show fit
|
0.01 |
ENST00000535011.6
ENST00000542996.6
ENST00000249364.9
ENST00000449187.6
|
CALU
|
calumenin
|
|
chr7_-_106661148
Show fit
|
0.01 |
ENST00000523505.3
|
CCDC71L
|
coiled-coil domain containing 71 like
|
|
chr1_-_169893876
Show fit
|
0.01 |
ENST00000367771.11
ENST00000367772.8
|
SCYL3
|
SCY1 like pseudokinase 3
|
|
chr12_-_31792290
Show fit
|
0.01 |
ENST00000340398.5
|
H3-5
|
H3.5 histone
|
|
chr20_-_35664837
Show fit
|
0.01 |
ENST00000414711.5
ENST00000416778.5
ENST00000397442.5
ENST00000440240.5
ENST00000412056.5
ENST00000352393.8
ENST00000458038.5
ENST00000397443.7
ENST00000420363.5
ENST00000434795.5
ENST00000437100.5
ENST00000414664.5
ENST00000359646.1
ENST00000424458.5
ENST00000374114.8
ENST00000374104.7
|
CPNE1
RBM12
|
copine 1
RNA binding motif protein 12
|
|
chr1_+_151070740
Show fit
|
0.01 |
ENST00000368918.8
|
GABPB2
|
GA binding protein transcription factor subunit beta 2
|
|
chr1_-_157045709
Show fit
|
0.01 |
ENST00000368194.8
|
ARHGEF11
|
Rho guanine nucleotide exchange factor 11
|
|
chr20_-_610299
Show fit
|
0.00 |
ENST00000246080.4
|
TCF15
|
transcription factor 15
|
|
chr3_+_180912656
Show fit
|
0.00 |
ENST00000357559.9
ENST00000491062.5
ENST00000468861.5
ENST00000445140.6
ENST00000484958.5
|
FXR1
|
FMR1 autosomal homolog 1
|
|
chr1_-_37859583
Show fit
|
0.00 |
ENST00000373036.5
|
MTF1
|
metal regulatory transcription factor 1
|
|
chr18_-_47930630
Show fit
|
0.00 |
ENST00000262160.11
|
SMAD2
|
SMAD family member 2
|
|
chr1_+_107141022
Show fit
|
0.00 |
ENST00000370067.5
ENST00000370068.6
|
NTNG1
|
netrin G1
|
|
chr19_+_18340629
Show fit
|
0.00 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I
|
|
chr1_+_244653081
Show fit
|
0.00 |
ENST00000263831.11
ENST00000302550.16
|
DESI2
|
desumoylating isopeptidase 2
|
|
chr15_+_50424377
Show fit
|
0.00 |
ENST00000560297.5
ENST00000396444.7
ENST00000425032.7
ENST00000307179.9
ENST00000625664.2
|
USP8
|
ubiquitin specific peptidase 8
|
|
chr9_-_10612966
Show fit
|
0.00 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D
|
|
chr6_-_62286161
Show fit
|
0.00 |
ENST00000281156.5
|
KHDRBS2
|
KH RNA binding domain containing, signal transduction associated 2
|
|
chr15_+_41417104
Show fit
|
0.00 |
ENST00000389629.8
|
RTF1
|
RTF1 homolog, Paf1/RNA polymerase II complex component
|
|
chr12_-_120250145
Show fit
|
0.00 |
ENST00000458477.6
|
PXN
|
paxillin
|
|
chr3_+_14125042
Show fit
|
0.00 |
ENST00000306077.5
|
TMEM43
|
transmembrane protein 43
|
|
chr9_-_3525968
Show fit
|
0.00 |
ENST00000382004.7
ENST00000617270.5
ENST00000449190.5
|
RFX3
|
regulatory factor X3
|
|
chr1_-_93180261
Show fit
|
0.00 |
ENST00000370280.1
ENST00000479918.5
|
TMED5
|
transmembrane p24 trafficking protein 5
|
|
chr7_+_141074038
Show fit
|
0.00 |
ENST00000565468.6
ENST00000610315.1
|
TMEM178B
|
transmembrane protein 178B
|
|
chr3_+_169966764
Show fit
|
0.00 |
ENST00000337002.9
ENST00000480708.5
|
SEC62
|
SEC62 homolog, preprotein translocation factor
|