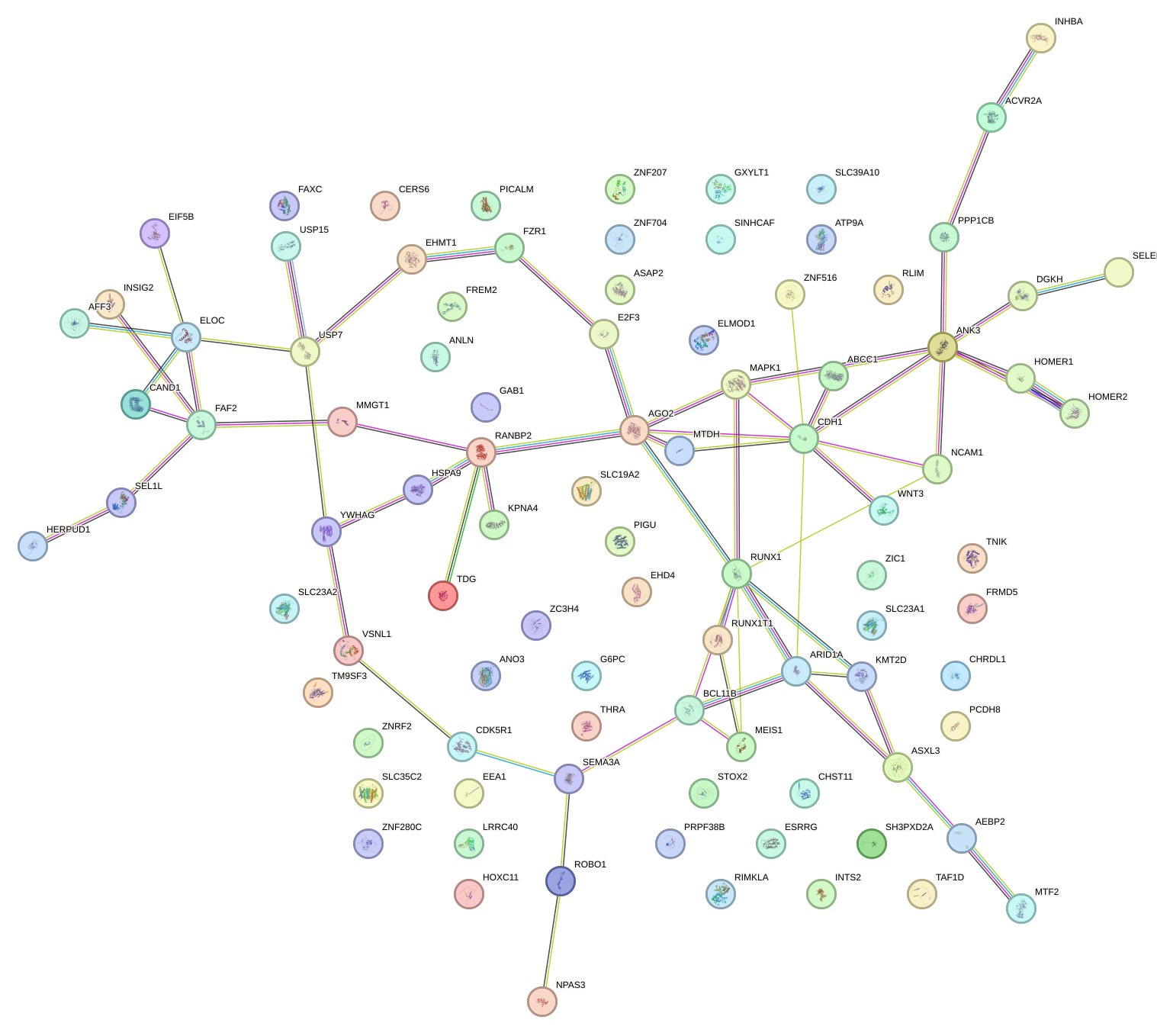
|
chr10_-_103855406
Show fit
|
0.10 |
ENST00000355946.6
ENST00000369774.8
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr2_+_17541157
Show fit
|
0.09 |
ENST00000406397.1
|
VSNL1
|
visinin like 1
|
|
chr14_-_99272184
Show fit
|
0.08 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B
|
|
chr7_-_84194781
Show fit
|
0.07 |
ENST00000265362.9
|
SEMA3A
|
semaphorin 3A
|
|
chr7_-_41703062
Show fit
|
0.07 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A
|
|
chr16_+_68737284
Show fit
|
0.07 |
ENST00000261769.10
ENST00000422392.6
|
CDH1
|
cadherin 1
|
|
chr6_-_99349647
Show fit
|
0.05 |
ENST00000389677.6
|
FAXC
|
failed axon connections homolog, metaxin like GST domain containing
|
|
chr12_-_31326111
Show fit
|
0.05 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor
|
|
chr12_+_104456962
Show fit
|
0.05 |
ENST00000547956.1
ENST00000549260.5
|
CHST11
|
carbohydrate sulfotransferase 11
|
|
chr7_+_36389852
Show fit
|
0.04 |
ENST00000265748.7
|
ANLN
|
anillin actin binding protein
|
|
chr1_-_217089627
Show fit
|
0.04 |
ENST00000361525.7
|
ESRRG
|
estrogen related receptor gamma
|
|
chr15_-_44194407
Show fit
|
0.04 |
ENST00000484674.5
|
FRMD5
|
FERM domain containing 5
|
|
chrX_-_130268883
Show fit
|
0.04 |
ENST00000447817.1
ENST00000370978.9
|
ZNF280C
|
zinc finger protein 280C
|
|
chr17_-_46818680
Show fit
|
0.03 |
ENST00000225512.6
|
WNT3
|
Wnt family member 3
|
|
chr8_+_103819244
Show fit
|
0.03 |
ENST00000262231.14
ENST00000507740.5
ENST00000408894.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr8_-_140635617
Show fit
|
0.03 |
ENST00000220592.10
|
AGO2
|
argonaute RISC catalytic component 2
|
|
chr2_+_26346086
Show fit
|
0.03 |
ENST00000613142.4
ENST00000260585.12
ENST00000447170.1
|
SELENOI
|
selenoprotein I
|
|
chr9_-_23821275
Show fit
|
0.03 |
ENST00000380110.8
|
ELAVL2
|
ELAV like RNA binding protein 2
|
|
chr2_+_99337364
Show fit
|
0.03 |
ENST00000617677.1
ENST00000289371.11
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr15_-_82952683
Show fit
|
0.03 |
ENST00000450735.7
ENST00000304231.12
|
HOMER2
|
homer scaffold protein 2
|
|
chr2_+_108719473
Show fit
|
0.03 |
ENST00000283195.11
|
RANBP2
|
RAN binding protein 2
|
|
chr9_-_123184233
Show fit
|
0.02 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr17_+_32486975
Show fit
|
0.02 |
ENST00000313401.4
|
CDK5R1
|
cyclin dependent kinase 5 regulatory subunit 1
|
|
chr3_-_79019444
Show fit
|
0.02 |
ENST00000618833.4
ENST00000436010.6
ENST00000618846.4
|
ROBO1
|
roundabout guidance receptor 1
|
|
chr20_-_5001474
Show fit
|
0.02 |
ENST00000338244.6
|
SLC23A2
|
solute carrier family 23 member 2
|
|
chr13_+_42048645
Show fit
|
0.02 |
ENST00000337343.9
ENST00000261491.9
ENST00000611224.1
|
DGKH
|
diacylglycerol kinase eta
|
|
chr2_+_118088432
Show fit
|
0.02 |
ENST00000245787.9
|
INSIG2
|
insulin induced gene 2
|
|
chr16_+_7332839
Show fit
|
0.02 |
ENST00000355637.9
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr1_+_93079264
Show fit
|
0.02 |
ENST00000370298.9
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr9_-_101487091
Show fit
|
0.02 |
ENST00000374847.5
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4
|
|
chr2_+_195656734
Show fit
|
0.02 |
ENST00000409086.7
|
SLC39A10
|
solute carrier family 39 member 10
|
|
chr16_-_8963870
Show fit
|
0.02 |
ENST00000344836.9
|
USP7
|
ubiquitin specific peptidase 7
|
|
chr12_-_92929236
Show fit
|
0.02 |
ENST00000322349.13
|
EEA1
|
early endosome antigen 1
|
|
chr1_+_26695993
Show fit
|
0.02 |
ENST00000324856.13
ENST00000637465.1
|
ARID1A
|
AT-rich interaction domain 1A
|
|
chr5_-_79512794
Show fit
|
0.02 |
ENST00000282260.10
ENST00000508576.5
ENST00000535690.1
|
HOMER1
|
homer scaffold protein 1
|
|
chr14_+_32939243
Show fit
|
0.02 |
ENST00000346562.6
ENST00000548645.5
ENST00000356141.8
ENST00000357798.9
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chrX_-_74614612
Show fit
|
0.02 |
ENST00000349225.2
ENST00000332687.11
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr1_-_169485931
Show fit
|
0.01 |
ENST00000367804.4
ENST00000646596.1
ENST00000236137.10
|
SLC19A2
|
solute carrier family 19 member 2
|
|
chr20_-_46363174
Show fit
|
0.01 |
ENST00000372227.5
|
SLC35C2
|
solute carrier family 35 member C2
|
|
chr2_+_147845020
Show fit
|
0.01 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A
|
|
chr20_-_51768327
Show fit
|
0.01 |
ENST00000311637.9
ENST00000338821.6
|
ATP9A
|
ATPase phospholipid transporting 9A (putative)
|
|
chr21_-_34888683
Show fit
|
0.01 |
ENST00000344691.8
ENST00000358356.9
|
RUNX1
|
RUNX family transcription factor 1
|
|
chr11_-_86069043
Show fit
|
0.01 |
ENST00000532317.5
ENST00000528256.1
ENST00000393346.8
ENST00000526033.5
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_+_67269328
Show fit
|
0.01 |
ENST00000545606.6
|
CAND1
|
cullin associated and neddylation dissociated 1
|
|
chr10_-_96586975
Show fit
|
0.01 |
ENST00000371142.9
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr3_+_147409357
Show fit
|
0.01 |
ENST00000282928.5
|
ZIC1
|
Zic family member 1
|
|
chr17_-_61928005
Show fit
|
0.01 |
ENST00000444766.7
|
INTS2
|
integrator complex subunit 2
|
|
chr16_+_15949549
Show fit
|
0.01 |
ENST00000399408.7
ENST00000572882.3
ENST00000677164.1
|
ABCC1
|
ATP binding cassette subfamily C member 1
|
|
chr8_+_97644164
Show fit
|
0.01 |
ENST00000336273.8
|
MTDH
|
metadherin
|
|
chr2_-_100104530
Show fit
|
0.01 |
ENST00000432037.5
ENST00000673232.1
ENST00000423966.6
ENST00000409236.6
|
AFF3
|
AF4/FMR2 family member 3
|
|
chr1_+_108692289
Show fit
|
0.01 |
ENST00000370025.9
ENST00000370022.9
ENST00000370021.1
|
PRPF38B
|
pre-mRNA processing factor 38B
|
|
chr10_-_60389833
Show fit
|
0.01 |
ENST00000280772.7
|
ANK3
|
ankyrin 3
|
|
chr2_+_168456215
Show fit
|
0.01 |
ENST00000392687.4
ENST00000305747.11
|
CERS6
|
ceramide synthase 6
|
|
chr8_-_73972276
Show fit
|
0.01 |
ENST00000518127.5
|
ELOC
|
elongin C
|
|
chr13_-_52848632
Show fit
|
0.01 |
ENST00000377942.7
ENST00000338862.5
|
PCDH8
|
protocadherin 8
|
|
chr17_+_40062956
Show fit
|
0.01 |
ENST00000450525.7
|
THRA
|
thyroid hormone receptor alpha
|
|
chr11_+_107591077
Show fit
|
0.01 |
ENST00000531234.5
ENST00000265840.12
|
ELMOD1
|
ELMO domain containing 1
|
|
chr9_+_137618986
Show fit
|
0.01 |
ENST00000462484.5
ENST00000630754.2
ENST00000460843.6
ENST00000626216.2
ENST00000629335.2
|
EHMT1
|
euchromatic histone lysine methyltransferase 1
|
|
chrX_-_135973975
Show fit
|
0.01 |
ENST00000305963.3
ENST00000680510.1
ENST00000679621.1
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr6_+_110874775
Show fit
|
0.01 |
ENST00000675380.1
ENST00000368882.8
ENST00000368877.9
ENST00000368885.8
ENST00000672937.2
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr11_-_93741479
Show fit
|
0.01 |
ENST00000448108.7
ENST00000532455.1
|
TAF1D
|
TATA-box binding protein associated factor, RNA polymerase I subunit D
|
|
chr3_-_160565560
Show fit
|
0.01 |
ENST00000334256.9
ENST00000676866.1
ENST00000469804.1
|
KPNA4
|
karyopherin subunit alpha 4
|
|
chr18_+_33578213
Show fit
|
0.01 |
ENST00000681521.1
ENST00000269197.12
|
ASXL3
|
ASXL transcriptional regulator 3
|
|
chr15_-_41972504
Show fit
|
0.01 |
ENST00000220325.9
|
EHD4
|
EH domain containing 4
|
|
chr12_+_62260338
Show fit
|
0.01 |
ENST00000353364.7
ENST00000549523.5
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr3_-_171460368
Show fit
|
0.01 |
ENST00000436636.7
ENST00000465393.1
ENST00000341852.10
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr7_-_76358982
Show fit
|
0.01 |
ENST00000307630.5
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma
|
|
chr6_+_20401864
Show fit
|
0.01 |
ENST00000346618.8
ENST00000613242.4
|
E2F3
|
E2F transcription factor 3
|
|
chr7_+_30284574
Show fit
|
0.01 |
ENST00000323037.5
|
ZNRF2
|
zinc and ring finger 2
|
|
chr4_+_183905266
Show fit
|
0.01 |
ENST00000308497.9
|
STOX2
|
storkhead box 2
|
|
chr1_+_42380772
Show fit
|
0.01 |
ENST00000431473.4
ENST00000410070.6
|
RIMKLA
|
ribosomal modification protein rimK like family member A
|
|
chr12_-_42144823
Show fit
|
0.01 |
ENST00000398675.8
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr11_+_112961402
Show fit
|
0.01 |
ENST00000613217.4
ENST00000316851.12
ENST00000620046.4
ENST00000531044.5
ENST00000529356.5
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr12_-_49060742
Show fit
|
0.01 |
ENST00000301067.12
ENST00000683543.1
|
KMT2D
|
lysine methyltransferase 2D
|
|
chr12_+_103965863
Show fit
|
0.01 |
ENST00000392872.8
ENST00000537100.5
|
TDG
|
thymine DNA glycosylase
|
|
chr16_+_22008083
Show fit
|
0.01 |
ENST00000542527.7
ENST00000569656.5
ENST00000562695.1
|
MOSMO
|
modulator of smoothened
|
|
chr12_+_19439469
Show fit
|
0.01 |
ENST00000266508.14
|
AEBP2
|
AE binding protein 2
|
|
chr13_+_38687068
Show fit
|
0.01 |
ENST00000280481.9
|
FREM2
|
FRAS1 related extracellular matrix 2
|
|
chr5_+_176448363
Show fit
|
0.01 |
ENST00000261942.7
|
FAF2
|
Fas associated factor family member 2
|
|
chr4_+_143336762
Show fit
|
0.01 |
ENST00000262995.8
|
GAB1
|
GRB2 associated binding protein 1
|
|
chr2_+_66435558
Show fit
|
0.01 |
ENST00000488550.5
|
MEIS1
|
Meis homeobox 1
|
|
chr5_-_138575359
Show fit
|
0.01 |
ENST00000297185.9
ENST00000678300.1
ENST00000677425.1
ENST00000677064.1
ENST00000507115.6
|
HSPA9
|
heat shock protein family A (Hsp70) member 9
|
|
chr12_+_53973108
Show fit
|
0.01 |
ENST00000546378.1
ENST00000243082.4
|
HOXC11
|
homeobox C11
|
|
chr17_+_42900791
Show fit
|
0.01 |
ENST00000592383.5
ENST00000253801.7
ENST00000585489.1
|
G6PC1
|
glucose-6-phosphatase catalytic subunit 1
|
|
chr10_+_96832252
Show fit
|
0.01 |
ENST00000676187.1
ENST00000675687.1
ENST00000676123.1
ENST00000675471.1
ENST00000371103.8
ENST00000421806.4
ENST00000675250.1
ENST00000540664.6
ENST00000676414.1
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr16_+_56932134
Show fit
|
0.01 |
ENST00000439977.7
ENST00000300302.9
ENST00000344114.8
ENST00000379792.6
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1
|
|
chr7_-_112206380
Show fit
|
0.01 |
ENST00000437633.6
ENST00000428084.6
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr14_-_81533800
Show fit
|
0.01 |
ENST00000555824.5
ENST00000557372.1
ENST00000336735.9
|
SEL1L
|
SEL1L adaptor subunit of ERAD E3 ubiquitin ligase
|
|
chr11_+_26332117
Show fit
|
0.01 |
ENST00000531646.1
ENST00000256737.8
|
ANO3
|
anoctamin 3
|
|
chr1_-_70205531
Show fit
|
0.01 |
ENST00000370952.4
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr22_-_21867610
Show fit
|
0.01 |
ENST00000215832.11
ENST00000398822.7
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr8_-_80874771
Show fit
|
0.01 |
ENST00000327835.7
|
ZNF704
|
zinc finger protein 704
|
|
chrX_-_110795765
Show fit
|
0.01 |
ENST00000372045.5
ENST00000394797.8
ENST00000372042.6
ENST00000482160.5
ENST00000444321.2
|
CHRDL1
|
chordin like 1
|
|
chr18_-_76495191
Show fit
|
0.01 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516
|
|
chr2_+_9206762
Show fit
|
0.01 |
ENST00000315273.4
ENST00000281419.8
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr2_+_28751802
Show fit
|
0.01 |
ENST00000296122.10
ENST00000395366.3
|
PPP1CB
|
protein phosphatase 1 catalytic subunit beta
|
|
chr19_-_47113756
Show fit
|
0.01 |
ENST00000253048.10
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr17_+_32350132
Show fit
|
0.01 |
ENST00000321233.10
ENST00000394673.6
ENST00000394670.9
ENST00000579634.5
ENST00000580759.5
ENST00000342555.10
ENST00000577908.5
ENST00000394679.9
ENST00000582165.1
|
ZNF207
|
zinc finger protein 207
|
|
chr2_+_5692357
Show fit
|
0.00 |
ENST00000322002.5
|
SOX11
|
SRY-box transcription factor 11
|
|
chr17_+_31094969
Show fit
|
0.00 |
ENST00000431387.8
ENST00000358273.9
|
NF1
|
neurofibromin 1
|
|
chr2_+_234952009
Show fit
|
0.00 |
ENST00000392011.7
|
SH3BP4
|
SH3 domain binding protein 4
|
|
chr8_-_30083053
Show fit
|
0.00 |
ENST00000256255.11
ENST00000523761.1
|
SARAF
|
store-operated calcium entry associated regulatory factor
|
|
chr11_+_33257265
Show fit
|
0.00 |
ENST00000303296.9
ENST00000379016.7
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr4_-_23890035
Show fit
|
0.00 |
ENST00000507380.1
ENST00000264867.7
|
PPARGC1A
|
PPARG coactivator 1 alpha
|
|
chr4_+_71339014
Show fit
|
0.00 |
ENST00000340595.4
|
SLC4A4
|
solute carrier family 4 member 4
|
|
chr13_+_57631735
Show fit
|
0.00 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17
|
|
chr14_-_23551729
Show fit
|
0.00 |
ENST00000419474.5
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr2_-_23927107
Show fit
|
0.00 |
ENST00000238789.10
|
ATAD2B
|
ATPase family AAA domain containing 2B
|
|
chr2_-_29074515
Show fit
|
0.00 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator
|
|
chr15_+_44537136
Show fit
|
0.00 |
ENST00000261868.10
ENST00000535391.5
|
EIF3J
|
eukaryotic translation initiation factor 3 subunit J
|
|
chr9_-_72364504
Show fit
|
0.00 |
ENST00000237937.7
ENST00000343431.6
ENST00000376956.3
|
ZFAND5
|
zinc finger AN1-type containing 5
|
|
chr15_+_56918612
Show fit
|
0.00 |
ENST00000438423.6
ENST00000267811.9
ENST00000333725.10
ENST00000559609.5
|
TCF12
|
transcription factor 12
|
|
chr1_-_179229671
Show fit
|
0.00 |
ENST00000502732.6
ENST00000392043.4
|
ABL2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase
|
|
chr5_+_36876731
Show fit
|
0.00 |
ENST00000282516.13
ENST00000448238.2
|
NIPBL
|
NIPBL cohesin loading factor
|
|
chrX_+_71095838
Show fit
|
0.00 |
ENST00000374259.8
|
FOXO4
|
forkhead box O4
|
|
chr20_+_10218808
Show fit
|
0.00 |
ENST00000254976.7
ENST00000304886.6
|
SNAP25
|
synaptosome associated protein 25
|
|
chr6_+_35213948
Show fit
|
0.00 |
ENST00000274938.8
|
SCUBE3
|
signal peptide, CUB domain and EGF like domain containing 3
|
|
chr2_+_124025280
Show fit
|
0.00 |
ENST00000431078.1
ENST00000682447.1
|
CNTNAP5
|
contactin associated protein family member 5
|
|
chr2_+_227472132
Show fit
|
0.00 |
ENST00000409979.6
ENST00000310078.13
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr12_-_123533705
Show fit
|
0.00 |
ENST00000636882.1
ENST00000376874.9
|
RILPL1
|
Rab interacting lysosomal protein like 1
|
|
chr6_-_93419545
Show fit
|
0.00 |
ENST00000369297.1
ENST00000369303.9
ENST00000680224.1
ENST00000681532.1
ENST00000679565.1
|
EPHA7
|
EPH receptor A7
|
|
chr10_+_102918276
Show fit
|
0.00 |
ENST00000369878.9
ENST00000369875.3
|
CNNM2
|
cyclin and CBS domain divalent metal cation transport mediator 2
|
|
chr5_-_133968578
Show fit
|
0.00 |
ENST00000231512.5
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr2_+_104854104
Show fit
|
0.00 |
ENST00000361360.4
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr1_-_233295712
Show fit
|
0.00 |
ENST00000258229.14
|
PCNX2
|
pecanex 2
|
|
chr5_+_69093980
Show fit
|
0.00 |
ENST00000380860.8
ENST00000396591.8
ENST00000504103.5
ENST00000502979.1
|
SLC30A5
|
solute carrier family 30 member 5
|
|
chr4_-_65670339
Show fit
|
0.00 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5
|
|
chr1_-_35193135
Show fit
|
0.00 |
ENST00000357214.6
|
SFPQ
|
splicing factor proline and glutamine rich
|
|
chr7_-_26200734
Show fit
|
0.00 |
ENST00000354667.8
ENST00000618183.5
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr20_+_63895114
Show fit
|
0.00 |
ENST00000360864.9
|
DNAJC5
|
DnaJ heat shock protein family (Hsp40) member C5
|
|
chr2_+_190408324
Show fit
|
0.00 |
ENST00000417958.5
ENST00000432036.5
ENST00000392328.6
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr5_+_144205250
Show fit
|
0.00 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16
|
|
chr10_-_86521737
Show fit
|
0.00 |
ENST00000298767.10
|
WAPL
|
WAPL cohesin release factor
|
|
chr3_-_168095885
Show fit
|
0.00 |
ENST00000470487.6
|
GOLIM4
|
golgi integral membrane protein 4
|
|
chr3_-_101513175
Show fit
|
0.00 |
ENST00000394091.5
ENST00000394094.6
ENST00000394095.7
ENST00000348610.3
ENST00000314261.11
|
SENP7
|
SUMO specific peptidase 7
|
|
chr13_+_32586443
Show fit
|
0.00 |
ENST00000315596.15
|
PDS5B
|
PDS5 cohesin associated factor B
|
|
chr2_-_69643703
Show fit
|
0.00 |
ENST00000406297.7
ENST00000409085.9
|
AAK1
|
AP2 associated kinase 1
|
|
chr1_-_244451896
Show fit
|
0.00 |
ENST00000366535.4
|
ADSS2
|
adenylosuccinate synthase 2
|
|
chr5_+_151771884
Show fit
|
0.00 |
ENST00000627077.2
ENST00000678976.1
ENST00000677408.1
ENST00000678070.1
ENST00000678964.1
ENST00000678925.1
ENST00000394123.7
ENST00000522761.6
ENST00000676827.1
|
G3BP1
|
G3BP stress granule assembly factor 1
|
|
chr5_-_131635030
Show fit
|
0.00 |
ENST00000308008.10
ENST00000296859.10
ENST00000627212.2
ENST00000507093.5
ENST00000509018.6
ENST00000510071.5
|
RAPGEF6
|
Rap guanine nucleotide exchange factor 6
|
|
chr11_+_111602380
Show fit
|
0.00 |
ENST00000304987.4
|
SIK2
|
salt inducible kinase 2
|
|
chr17_-_18363504
Show fit
|
0.00 |
ENST00000583780.1
ENST00000316694.8
ENST00000352886.10
|
SHMT1
|
serine hydroxymethyltransferase 1
|
|
chr20_+_3471003
Show fit
|
0.00 |
ENST00000262919.10
ENST00000446916.2
|
ATRN
|
attractin
|
|
chr12_-_21941402
Show fit
|
0.00 |
ENST00000326684.8
ENST00000682068.1
ENST00000621589.2
ENST00000261200.9
ENST00000683676.1
|
ABCC9
|
ATP binding cassette subfamily C member 9
|
|
chr10_+_43138435
Show fit
|
0.00 |
ENST00000374466.4
|
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2
|
|
chr3_+_30606574
Show fit
|
0.00 |
ENST00000295754.10
ENST00000359013.4
|
TGFBR2
|
transforming growth factor beta receptor 2
|
|
chr5_+_62306228
Show fit
|
0.00 |
ENST00000381103.7
|
KIF2A
|
kinesin family member 2A
|
|
chr10_-_119596495
Show fit
|
0.00 |
ENST00000369093.6
|
TIAL1
|
TIA1 cytotoxic granule associated RNA binding protein like 1
|
|
chr3_+_113747022
Show fit
|
0.00 |
ENST00000273398.8
ENST00000496747.5
ENST00000475322.1
|
ATP6V1A
|
ATPase H+ transporting V1 subunit A
|
|
chr3_+_10992717
Show fit
|
0.00 |
ENST00000642767.1
ENST00000425938.6
ENST00000642515.1
ENST00000643498.1
ENST00000646072.1
ENST00000646570.1
ENST00000287766.10
ENST00000645281.1
ENST00000642820.1
ENST00000645054.1
ENST00000642735.1
ENST00000646022.1
ENST00000645776.1
ENST00000645592.1
ENST00000646924.1
ENST00000645974.1
ENST00000644314.1
ENST00000642639.1
ENST00000646060.1
ENST00000642201.1
ENST00000646487.1
ENST00000647194.1
ENST00000646088.1
|
SLC6A1
|
solute carrier family 6 member 1
|
|
chr16_-_53703883
Show fit
|
0.00 |
ENST00000262135.9
ENST00000564374.5
ENST00000566096.5
|
RPGRIP1L
|
RPGRIP1 like
|
|
chr18_+_57435366
Show fit
|
0.00 |
ENST00000491143.3
|
ONECUT2
|
one cut homeobox 2
|
|
chr1_-_243850070
Show fit
|
0.00 |
ENST00000366539.6
ENST00000672578.1
|
AKT3
|
AKT serine/threonine kinase 3
|
|
chr17_+_31936993
Show fit
|
0.00 |
ENST00000322652.10
|
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit
|
|
chr3_+_33798557
Show fit
|
0.00 |
ENST00000457054.6
ENST00000413073.1
ENST00000307296.8
|
PDCD6IP
|
programmed cell death 6 interacting protein
|
|
chr1_+_213987929
Show fit
|
0.00 |
ENST00000498508.6
ENST00000366958.9
|
PROX1
|
prospero homeobox 1
|
|
chr4_+_56907876
Show fit
|
0.00 |
ENST00000640168.2
ENST00000309042.12
|
REST
|
RE1 silencing transcription factor
|
|
chr3_-_53891836
Show fit
|
0.00 |
ENST00000495461.6
ENST00000541726.5
|
SELENOK
|
selenoprotein K
|
|
chr22_-_21952827
Show fit
|
0.00 |
ENST00000397495.8
ENST00000263212.10
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent 1F
|
|
chr4_-_42657085
Show fit
|
0.00 |
ENST00000264449.14
ENST00000510289.1
ENST00000381668.9
|
ATP8A1
|
ATPase phospholipid transporting 8A1
|
|
chr5_-_131796965
Show fit
|
0.00 |
ENST00000514667.1
ENST00000511848.1
ENST00000510461.6
|
ENSG00000273217.1
FNIP1
|
novel protein
folliculin interacting protein 1
|
|
chr7_+_103297425
Show fit
|
0.00 |
ENST00000428154.5
ENST00000249269.9
|
PMPCB
|
peptidase, mitochondrial processing subunit beta
|
|
chr17_-_62065248
Show fit
|
0.00 |
ENST00000397786.7
|
MED13
|
mediator complex subunit 13
|
|
chr3_+_173398438
Show fit
|
0.00 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1
|
|
chr5_-_168579319
Show fit
|
0.00 |
ENST00000522176.1
ENST00000239231.7
|
PANK3
|
pantothenate kinase 3
|
|
chr2_+_177212724
Show fit
|
0.00 |
ENST00000677043.1
ENST00000677863.1
ENST00000676681.1
ENST00000678111.1
ENST00000676874.1
ENST00000676736.1
ENST00000411529.6
ENST00000435711.5
|
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chr7_-_135510074
Show fit
|
0.00 |
ENST00000428680.6
ENST00000423368.6
ENST00000451834.5
ENST00000361528.8
ENST00000541284.5
ENST00000315544.6
|
CNOT4
|
CCR4-NOT transcription complex subunit 4
|
|
chr2_-_181680490
Show fit
|
0.00 |
ENST00000684145.1
ENST00000295108.4
ENST00000684079.1
ENST00000683430.1
|
CERKL
NEUROD1
|
ceramide kinase like
neuronal differentiation 1
|
|
chr14_+_100239121
Show fit
|
0.00 |
ENST00000262238.10
|
YY1
|
YY1 transcription factor
|
|
chr11_+_125592826
Show fit
|
0.00 |
ENST00000529196.5
ENST00000392708.9
ENST00000649491.1
ENST00000531491.5
|
STT3A
|
STT3 oligosaccharyltransferase complex catalytic subunit A
|