Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for AIRE
Z-value: 0.32
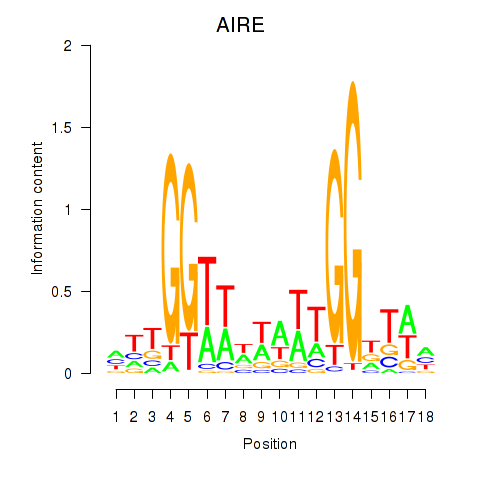
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.17 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg38_v1_chr21_+_44285869_44285884 | 0.01 | 9.8e-01 | Click! |
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_75480800 | 0.56 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr12_+_75481204 | 0.53 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1 |
| chrM_+_12329 | 0.38 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr12_+_75480745 | 0.37 |
ENST00000266659.8
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr14_+_75632610 | 0.25 |
ENST00000555027.1
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr2_-_224947030 | 0.23 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr2_-_189179754 | 0.23 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr2_-_165204042 | 0.19 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr17_-_40565459 | 0.19 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
C-C motif chemokine receptor 7 |
| chr8_+_103372388 | 0.19 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_-_237414127 | 0.19 |
ENST00000472056.5
|
COL6A3
|
collagen type VI alpha 3 chain |
| chr4_-_185775271 | 0.18 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_141392616 | 0.16 |
ENST00000398604.3
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr12_+_50924005 | 0.15 |
ENST00000550502.1
|
METTL7A
|
methyltransferase like 7A |
| chrX_-_101052087 | 0.15 |
ENST00000372936.4
ENST00000545398.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B |
| chrX_-_101052054 | 0.15 |
ENST00000372939.5
ENST00000372935.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B |
| chr12_+_12049855 | 0.13 |
ENST00000586576.5
ENST00000464885.6 |
BCL2L14
|
BCL2 like 14 |
| chr9_-_110999458 | 0.13 |
ENST00000374430.6
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr9_+_87497222 | 0.13 |
ENST00000358077.9
|
DAPK1
|
death associated protein kinase 1 |
| chr12_+_12070932 | 0.13 |
ENST00000308721.9
|
BCL2L14
|
BCL2 like 14 |
| chr12_+_12049840 | 0.13 |
ENST00000589718.5
|
BCL2L14
|
BCL2 like 14 |
| chr10_-_100081854 | 0.13 |
ENST00000370418.8
|
CPN1
|
carboxypeptidase N subunit 1 |
| chr1_+_209938207 | 0.13 |
ENST00000472886.5
|
SYT14
|
synaptotagmin 14 |
| chr12_+_54854505 | 0.13 |
ENST00000308796.11
ENST00000619042.1 |
MUCL1
|
mucin like 1 |
| chr1_+_209938169 | 0.12 |
ENST00000367019.5
ENST00000537238.5 ENST00000637265.1 |
SYT14
|
synaptotagmin 14 |
| chr12_+_12071396 | 0.12 |
ENST00000396367.5
ENST00000266434.8 |
BCL2L14
|
BCL2 like 14 |
| chr20_+_8789517 | 0.11 |
ENST00000437439.2
|
PLCB1
|
phospholipase C beta 1 |
| chr6_-_31546552 | 0.11 |
ENST00000303892.10
ENST00000376151.4 |
ATP6V1G2
|
ATPase H+ transporting V1 subunit G2 |
| chr12_+_20695553 | 0.11 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr16_-_47459130 | 0.11 |
ENST00000565940.6
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr2_-_43676406 | 0.10 |
ENST00000475092.4
|
C1GALT1C1L
|
C1GALT1 specific chaperone 1 like |
| chr19_-_52048561 | 0.10 |
ENST00000594154.5
ENST00000598745.5 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr7_+_149872955 | 0.10 |
ENST00000421974.7
ENST00000456496.7 |
ATP6V0E2
|
ATPase H+ transporting V0 subunit e2 |
| chr19_-_52048803 | 0.09 |
ENST00000221315.10
|
ZNF432
|
zinc finger protein 432 |
| chr20_+_13008919 | 0.09 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr1_+_174874434 | 0.09 |
ENST00000478442.5
ENST00000465412.5 |
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr3_-_121749704 | 0.09 |
ENST00000393667.7
ENST00000340645.9 ENST00000614479.4 |
GOLGB1
|
golgin B1 |
| chr12_-_11134644 | 0.09 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chrM_+_5824 | 0.09 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr16_-_89418292 | 0.09 |
ENST00000683497.1
ENST00000642443.1 ENST00000644784.1 ENST00000647238.1 |
ENSG00000288715.1
ANKRD11
|
novel protein ankyrin repeat domain 11 |
| chr1_+_78649818 | 0.09 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr1_+_15153698 | 0.09 |
ENST00000400796.7
ENST00000376008.3 ENST00000434578.6 |
TMEM51
|
transmembrane protein 51 |
| chr15_+_21651844 | 0.08 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr9_-_21482313 | 0.08 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr3_-_126558926 | 0.08 |
ENST00000318225.3
|
C3orf22
|
chromosome 3 open reading frame 22 |
| chr15_+_22094522 | 0.08 |
ENST00000328795.5
|
OR4N4
|
olfactory receptor family 4 subfamily N member 4 |
| chrM_+_4467 | 0.08 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr1_+_241652275 | 0.08 |
ENST00000366552.6
ENST00000437684.7 |
WDR64
|
WD repeat domain 64 |
| chr6_+_36871841 | 0.08 |
ENST00000359359.6
|
C6orf89
|
chromosome 6 open reading frame 89 |
| chr2_+_37950432 | 0.07 |
ENST00000407257.5
ENST00000417700.6 ENST00000234195.7 ENST00000442857.5 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr16_+_50025217 | 0.07 |
ENST00000427478.7
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr10_+_89283685 | 0.07 |
ENST00000638108.1
|
IFIT2
|
interferon induced protein with tetratricopeptide repeats 2 |
| chrX_-_140784366 | 0.06 |
ENST00000674533.1
|
CDR1
|
cerebellar degeneration related protein 1 |
| chr19_-_51028015 | 0.06 |
ENST00000319720.11
|
KLK11
|
kallikrein related peptidase 11 |
| chr9_-_92536031 | 0.06 |
ENST00000344604.9
ENST00000375540.5 |
ECM2
|
extracellular matrix protein 2 |
| chr6_-_149484965 | 0.06 |
ENST00000409806.8
|
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr19_+_15615211 | 0.06 |
ENST00000617645.4
ENST00000612078.5 |
CYP4F8
|
cytochrome P450 family 4 subfamily F member 8 |
| chr11_-_71821548 | 0.06 |
ENST00000525199.1
|
ZNF705E
|
zinc finger protein 705E |
| chr13_-_46182136 | 0.06 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr20_-_35147285 | 0.06 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr21_+_38272250 | 0.06 |
ENST00000398932.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr10_+_94089034 | 0.06 |
ENST00000676102.1
ENST00000371385.8 |
PLCE1
|
phospholipase C epsilon 1 |
| chr2_+_165294031 | 0.06 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr13_-_48413105 | 0.06 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr21_+_38272410 | 0.06 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr3_+_136819069 | 0.05 |
ENST00000393079.3
ENST00000446465.3 |
SLC35G2
|
solute carrier family 35 member G2 |
| chrM_+_7586 | 0.05 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr16_+_14708944 | 0.05 |
ENST00000526520.5
ENST00000531598.6 |
NPIPA3
|
nuclear pore complex interacting protein family member A3 |
| chr19_+_13151975 | 0.05 |
ENST00000588173.1
|
IER2
|
immediate early response 2 |
| chr17_+_63484840 | 0.05 |
ENST00000290863.10
ENST00000413513.7 |
ACE
|
angiotensin I converting enzyme |
| chr14_-_55191534 | 0.05 |
ENST00000395425.6
ENST00000247191.7 |
DLGAP5
|
DLG associated protein 5 |
| chr1_-_35031905 | 0.05 |
ENST00000317538.9
ENST00000357182.9 |
ZMYM6
|
zinc finger MYM-type containing 6 |
| chr11_+_7088991 | 0.05 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr19_+_44914702 | 0.05 |
ENST00000592885.5
ENST00000589781.1 |
APOC1
|
apolipoprotein C1 |
| chr11_-_117098415 | 0.05 |
ENST00000445177.6
ENST00000375300.6 ENST00000446921.6 |
SIK3
|
SIK family kinase 3 |
| chr19_-_51027954 | 0.05 |
ENST00000391804.7
|
KLK11
|
kallikrein related peptidase 11 |
| chr16_+_32066065 | 0.05 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr14_+_22112280 | 0.05 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr19_+_55769118 | 0.04 |
ENST00000341750.5
|
RFPL4AL1
|
ret finger protein like 4A like 1 |
| chr17_-_43942472 | 0.04 |
ENST00000225992.8
|
PPY
|
pancreatic polypeptide |
| chr14_-_24442662 | 0.04 |
ENST00000554698.5
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U member 1 |
| chr2_-_222656067 | 0.04 |
ENST00000281828.8
|
FARSB
|
phenylalanyl-tRNA synthetase subunit beta |
| chr1_-_21783134 | 0.04 |
ENST00000308271.14
|
USP48
|
ubiquitin specific peptidase 48 |
| chr6_-_26043704 | 0.04 |
ENST00000615966.2
|
H2BC3
|
H2B clustered histone 3 |
| chr19_+_44914588 | 0.04 |
ENST00000592535.6
|
APOC1
|
apolipoprotein C1 |
| chr7_-_15561986 | 0.04 |
ENST00000342526.8
|
AGMO
|
alkylglycerol monooxygenase |
| chr19_-_10380558 | 0.04 |
ENST00000524462.5
ENST00000525621.6 ENST00000531836.5 |
TYK2
|
tyrosine kinase 2 |
| chr17_+_3420568 | 0.04 |
ENST00000574571.4
|
OR3A3
|
olfactory receptor family 3 subfamily A member 3 |
| chr16_-_28491996 | 0.04 |
ENST00000568497.6
ENST00000357857.14 ENST00000395653.9 ENST00000635973.1 ENST00000568443.2 ENST00000568558.6 ENST00000565688.5 ENST00000637100.1 ENST00000359984.12 ENST00000636147.2 ENST00000567963.6 |
CLN3
|
CLN3 lysosomal/endosomal transmembrane protein, battenin |
| chr19_-_58573555 | 0.03 |
ENST00000599369.5
|
MZF1
|
myeloid zinc finger 1 |
| chr1_+_47137435 | 0.03 |
ENST00000371891.8
ENST00000371890.7 ENST00000619754.4 ENST00000294337.7 ENST00000620131.1 |
CYP4A22
|
cytochrome P450 family 4 subfamily A member 22 |
| chr19_+_44914833 | 0.03 |
ENST00000589078.1
ENST00000586638.5 |
APOC1
|
apolipoprotein C1 |
| chr2_+_113437691 | 0.03 |
ENST00000259199.9
ENST00000416503.6 ENST00000433343.6 |
CBWD2
|
COBW domain containing 2 |
| chr17_-_3398410 | 0.03 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor family 1 subfamily E member 1 |
| chr11_-_85665077 | 0.03 |
ENST00000527447.2
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr12_-_6470667 | 0.03 |
ENST00000361716.8
ENST00000396308.4 |
VAMP1
|
vesicle associated membrane protein 1 |
| chr2_-_37671633 | 0.03 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr4_-_65670478 | 0.03 |
ENST00000613740.5
ENST00000622150.4 ENST00000511294.1 |
EPHA5
|
EPH receptor A5 |
| chr9_+_105662457 | 0.03 |
ENST00000334077.6
|
TAL2
|
TAL bHLH transcription factor 2 |
| chr3_+_39383337 | 0.03 |
ENST00000650617.1
ENST00000431510.1 ENST00000645630.1 |
SLC25A38
|
solute carrier family 25 member 38 |
| chr3_-_19934189 | 0.03 |
ENST00000295824.14
|
EFHB
|
EF-hand domain family member B |
| chr17_+_79034185 | 0.03 |
ENST00000581774.5
|
C1QTNF1
|
C1q and TNF related 1 |
| chr3_+_189789672 | 0.03 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr9_-_19049319 | 0.03 |
ENST00000542071.2
ENST00000649457.1 |
SAXO1
|
stabilizer of axonemal microtubules 1 |
| chr2_-_240820945 | 0.03 |
ENST00000428768.2
ENST00000650053.1 ENST00000650130.1 |
KIF1A
|
kinesin family member 1A |
| chr4_-_174829212 | 0.03 |
ENST00000340217.5
ENST00000274093.8 |
GLRA3
|
glycine receptor alpha 3 |
| chr7_-_99735093 | 0.03 |
ENST00000611620.4
ENST00000620220.6 ENST00000336374.4 |
CYP3A7-CYP3A51P
CYP3A7
|
CYP3A7-CYP3A51P readthrough cytochrome P450 family 3 subfamily A member 7 |
| chr1_-_85708382 | 0.03 |
ENST00000370574.4
ENST00000431532.6 |
ZNHIT6
|
zinc finger HIT-type containing 6 |
| chr9_-_4666347 | 0.03 |
ENST00000381890.9
ENST00000682582.1 |
SPATA6L
|
spermatogenesis associated 6 like |
| chr3_-_36739791 | 0.03 |
ENST00000416516.2
|
DCLK3
|
doublecortin like kinase 3 |
| chr5_+_141245384 | 0.02 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chrM_-_14669 | 0.02 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 |
| chr12_-_75209701 | 0.02 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr2_-_27370009 | 0.02 |
ENST00000451130.6
|
EIF2B4
|
eukaryotic translation initiation factor 2B subunit delta |
| chr2_-_135876382 | 0.02 |
ENST00000264156.3
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr5_-_78549151 | 0.02 |
ENST00000515007.6
|
LHFPL2
|
LHFPL tetraspan subfamily member 2 |
| chr4_-_65670339 | 0.02 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5 |
| chrX_-_49080066 | 0.02 |
ENST00000634944.1
ENST00000423215.3 ENST00000465382.6 |
WDR45
|
WD repeat domain 45 |
| chr19_-_43883964 | 0.02 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr8_+_103298433 | 0.02 |
ENST00000522566.5
|
FZD6
|
frizzled class receptor 6 |
| chr19_-_23687163 | 0.02 |
ENST00000601010.5
ENST00000601935.5 ENST00000600313.5 ENST00000596211.5 ENST00000359788.9 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr19_-_3600581 | 0.02 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr21_+_38272291 | 0.02 |
ENST00000438657.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr14_-_24442765 | 0.02 |
ENST00000555365.5
ENST00000399395.8 ENST00000553930.5 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U member 1 |
| chr8_-_16186270 | 0.02 |
ENST00000445506.6
|
MSR1
|
macrophage scavenger receptor 1 |
| chr11_-_5324297 | 0.02 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2 |
| chr3_-_88149815 | 0.02 |
ENST00000467332.1
ENST00000462901.5 |
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr11_+_6926417 | 0.02 |
ENST00000610573.4
ENST00000278319.10 |
ZNF215
|
zinc finger protein 215 |
| chr1_-_51878711 | 0.02 |
ENST00000352171.12
|
NRDC
|
nardilysin convertase |
| chr4_-_146945841 | 0.02 |
ENST00000325106.9
|
TTC29
|
tetratricopeptide repeat domain 29 |
| chr17_+_7306975 | 0.02 |
ENST00000336452.11
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr13_+_52455429 | 0.02 |
ENST00000468284.1
ENST00000378034.7 ENST00000378037.9 ENST00000258607.10 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr12_-_6470643 | 0.02 |
ENST00000535180.5
ENST00000400911.7 |
VAMP1
|
vesicle associated membrane protein 1 |
| chr1_-_51878799 | 0.02 |
ENST00000354831.11
ENST00000544028.5 |
NRDC
|
nardilysin convertase |
| chr9_-_29739 | 0.02 |
ENST00000442898.5
|
WASHC1
|
WASH complex subunit 1 |
| chr19_+_55857437 | 0.02 |
ENST00000587891.5
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chr4_-_146945873 | 0.02 |
ENST00000502319.1
ENST00000504425.5 |
TTC29
|
tetratricopeptide repeat domain 29 |
| chr11_+_35189964 | 0.02 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr8_-_7452365 | 0.02 |
ENST00000458665.5
ENST00000528168.3 |
SPAG11B
|
sperm associated antigen 11B |
| chr4_-_146945807 | 0.02 |
ENST00000513335.5
|
TTC29
|
tetratricopeptide repeat domain 29 |
| chr8_+_53851786 | 0.02 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr2_-_165953750 | 0.02 |
ENST00000243344.8
ENST00000679799.1 ENST00000679840.1 ENST00000681606.1 ENST00000680448.1 |
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr1_+_207496147 | 0.02 |
ENST00000400960.7
ENST00000367049.9 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr4_-_103099811 | 0.01 |
ENST00000504285.5
ENST00000296424.9 |
BDH2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr17_+_56153458 | 0.01 |
ENST00000318698.6
ENST00000682825.1 ENST00000566473.6 |
ANKFN1
|
ankyrin repeat and fibronectin type III domain containing 1 |
| chr7_-_88306880 | 0.01 |
ENST00000414498.1
ENST00000301959.9 ENST00000380079.9 |
STEAP4
|
STEAP4 metalloreductase |
| chr3_+_88149947 | 0.01 |
ENST00000318887.8
ENST00000486971.1 |
C3orf38
|
chromosome 3 open reading frame 38 |
| chr19_+_16496383 | 0.01 |
ENST00000594035.5
ENST00000221671.8 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr17_+_43780425 | 0.01 |
ENST00000449302.8
|
CFAP97D1
|
CFAP97 domain containing 1 |
| chr12_-_11310420 | 0.01 |
ENST00000621732.4
ENST00000445719.2 ENST00000279575.7 |
PRB4
|
proline rich protein BstNI subfamily 4 |
| chr12_+_4649097 | 0.01 |
ENST00000648836.1
ENST00000266544.10 |
ENSG00000255639.3
NDUFA9
|
novel protein NADH:ubiquinone oxidoreductase subunit A9 |
| chr14_+_45135917 | 0.01 |
ENST00000267430.10
ENST00000556036.5 ENST00000542564.6 |
FANCM
|
FA complementation group M |
| chr14_+_22105305 | 0.01 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr6_+_28267044 | 0.01 |
ENST00000316606.10
|
ZSCAN26
|
zinc finger and SCAN domain containing 26 |
| chr6_+_46129930 | 0.01 |
ENST00000321037.5
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr12_-_62192762 | 0.01 |
ENST00000416284.8
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr1_+_100345018 | 0.01 |
ENST00000635056.2
ENST00000647005.1 |
CDC14A
|
cell division cycle 14A |
| chr1_+_111473972 | 0.01 |
ENST00000369718.4
|
C1orf162
|
chromosome 1 open reading frame 162 |
| chr19_-_16496156 | 0.00 |
ENST00000269881.8
|
CALR3
|
calreticulin 3 |
| chr1_-_16431371 | 0.00 |
ENST00000612240.1
|
SPATA21
|
spermatogenesis associated 21 |
| chr22_+_24806265 | 0.00 |
ENST00000400359.4
|
SGSM1
|
small G protein signaling modulator 1 |
| chr4_-_16084002 | 0.00 |
ENST00000447510.7
|
PROM1
|
prominin 1 |
| chr16_+_3204247 | 0.00 |
ENST00000304646.2
|
OR1F1
|
olfactory receptor family 1 subfamily F member 1 |
| chr1_+_162497805 | 0.00 |
ENST00000538489.5
ENST00000489294.2 |
UHMK1
|
U2AF homology motif kinase 1 |
| chr12_-_11269696 | 0.00 |
ENST00000381842.7
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr1_-_154627576 | 0.00 |
ENST00000648311.1
|
ADAR
|
adenosine deaminase RNA specific |
| chr22_+_41621977 | 0.00 |
ENST00000405506.2
|
XRCC6
|
X-ray repair cross complementing 6 |
| chr7_+_20647343 | 0.00 |
ENST00000443026.6
ENST00000406935.5 |
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chrX_-_103686687 | 0.00 |
ENST00000441076.7
ENST00000422355.5 ENST00000442614.5 ENST00000451301.5 |
MORF4L2
|
mortality factor 4 like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.0 | GO:0036233 | glycine import(GO:0036233) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 1.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0008241 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |