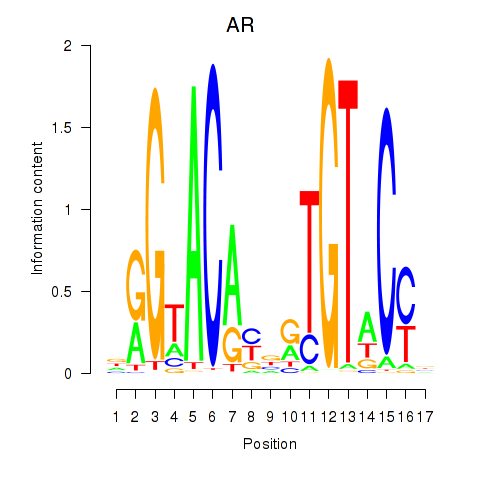
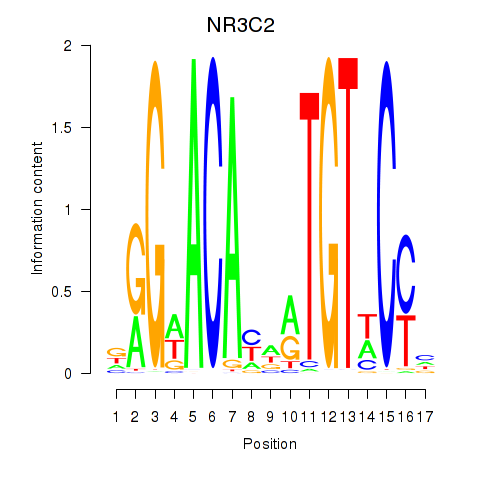
|
chr17_-_19386785
Show fit
|
1.21 |
ENST00000497081.6
|
MFAP4
|
microfibril associated protein 4
|
|
chr17_-_19387170
Show fit
|
1.16 |
ENST00000395592.6
ENST00000299610.5
|
MFAP4
|
microfibril associated protein 4
|
|
chr12_+_53050179
Show fit
|
0.66 |
ENST00000546602.5
ENST00000552570.5
ENST00000549700.5
|
TNS2
|
tensin 2
|
|
chr12_+_53050014
Show fit
|
0.66 |
ENST00000314250.11
|
TNS2
|
tensin 2
|
|
chr16_+_56632651
Show fit
|
0.56 |
ENST00000379818.4
ENST00000570233.1
|
MT1M
|
metallothionein 1M
|
|
chr3_-_58577367
Show fit
|
0.54 |
ENST00000464064.5
ENST00000360997.7
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr3_-_58577648
Show fit
|
0.49 |
ENST00000394481.5
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr1_+_22653189
Show fit
|
0.46 |
ENST00000432749.6
|
C1QB
|
complement C1q B chain
|
|
chr8_+_90001448
Show fit
|
0.39 |
ENST00000519410.5
ENST00000522161.5
ENST00000220764.7
ENST00000517761.5
ENST00000520227.1
|
DECR1
|
2,4-dienoyl-CoA reductase 1
|
|
chr19_+_35140022
Show fit
|
0.30 |
ENST00000588081.5
ENST00000589121.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr19_+_35139724
Show fit
|
0.29 |
ENST00000588715.5
ENST00000588607.5
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr15_+_31366138
Show fit
|
0.28 |
ENST00000558844.1
|
KLF13
|
Kruppel like factor 13
|
|
chr7_+_94656325
Show fit
|
0.28 |
ENST00000482108.1
ENST00000488574.5
ENST00000612748.1
ENST00000613043.1
|
PEG10
|
paternally expressed 10
|
|
chr16_+_67173935
Show fit
|
0.26 |
ENST00000566871.5
|
NOL3
|
nucleolar protein 3
|
|
chr16_+_56682461
Show fit
|
0.26 |
ENST00000562939.1
ENST00000394485.5
ENST00000567563.1
|
MT1X
ENSG00000259827.1
|
metallothionein 1X
novel transcript
|
|
chr1_+_22653228
Show fit
|
0.25 |
ENST00000509305.6
|
C1QB
|
complement C1q B chain
|
|
chr19_+_35139440
Show fit
|
0.24 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr5_-_142012973
Show fit
|
0.24 |
ENST00000503794.5
ENST00000510194.5
ENST00000504424.1
ENST00000513454.5
ENST00000311337.11
ENST00000503229.5
ENST00000500692.6
ENST00000504139.5
ENST00000505689.5
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr1_-_1214146
Show fit
|
0.21 |
ENST00000379236.4
|
TNFRSF4
|
TNF receptor superfamily member 4
|
|
chr22_+_22704265
Show fit
|
0.21 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22
|
|
chr16_+_67173971
Show fit
|
0.20 |
ENST00000563258.1
ENST00000568146.1
|
NOL3
|
nucleolar protein 3
|
|
chr16_+_56625775
Show fit
|
0.19 |
ENST00000330439.7
ENST00000568293.1
|
MT1E
|
metallothionein 1E
|
|
chr2_+_237487239
Show fit
|
0.19 |
ENST00000338530.8
ENST00000264605.8
ENST00000409373.5
|
MLPH
|
melanophilin
|
|
chr9_+_128920966
Show fit
|
0.17 |
ENST00000428610.5
ENST00000372592.8
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr3_+_52245721
Show fit
|
0.17 |
ENST00000323588.9
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent 1M
|
|
chr8_-_71844402
Show fit
|
0.15 |
ENST00000325509.5
|
MSC
|
musculin
|
|
chr20_+_15196834
Show fit
|
0.15 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2
|
|
chr11_-_236326
Show fit
|
0.14 |
ENST00000525237.1
ENST00000382743.9
ENST00000532956.5
ENST00000525319.5
ENST00000524564.5
|
SIRT3
|
sirtuin 3
|
|
chr13_+_113759219
Show fit
|
0.14 |
ENST00000375353.5
ENST00000488362.5
|
TMEM255B
|
transmembrane protein 255B
|
|
chr7_+_20615653
Show fit
|
0.13 |
ENST00000404938.7
|
ABCB5
|
ATP binding cassette subfamily B member 5
|
|
chr1_+_111619751
Show fit
|
0.13 |
ENST00000433097.5
ENST00000369709.3
|
RAP1A
|
RAP1A, member of RAS oncogene family
|
|
chr12_+_66189178
Show fit
|
0.13 |
ENST00000545837.1
|
IRAK3
|
interleukin 1 receptor associated kinase 3
|
|
chr12_-_56221701
Show fit
|
0.12 |
ENST00000615206.4
ENST00000549038.5
ENST00000552244.5
|
RNF41
|
ring finger protein 41
|
|
chr8_+_61287950
Show fit
|
0.12 |
ENST00000519846.5
ENST00000325897.5
ENST00000523868.2
ENST00000518592.5
|
CLVS1
|
clavesin 1
|
|
chr4_-_68951763
Show fit
|
0.12 |
ENST00000251566.9
|
UGT2A3
|
UDP glucuronosyltransferase family 2 member A3
|
|
chr16_-_66550142
Show fit
|
0.12 |
ENST00000417693.8
ENST00000299697.12
ENST00000451102.7
|
TK2
|
thymidine kinase 2
|
|
chr18_-_50819982
Show fit
|
0.12 |
ENST00000398439.8
ENST00000431965.6
ENST00000436348.6
|
MRO
|
maestro
|
|
chr3_-_15341368
Show fit
|
0.11 |
ENST00000408919.7
|
SH3BP5
|
SH3 domain binding protein 5
|
|
chr17_-_8152380
Show fit
|
0.11 |
ENST00000317276.9
|
PER1
|
period circadian regulator 1
|
|
chr14_-_106675544
Show fit
|
0.11 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66
|
|
chr17_+_39667964
Show fit
|
0.11 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase
|
|
chrY_-_13986473
Show fit
|
0.11 |
ENST00000250825.5
|
VCY
|
variable charge Y-linked
|
|
chr12_+_50925007
Show fit
|
0.11 |
ENST00000332160.5
|
METTL7A
|
methyltransferase like 7A
|
|
chrY_+_14056226
Show fit
|
0.10 |
ENST00000250823.5
|
VCY1B
|
variable charge Y-linked 1B
|
|
chr12_-_56221909
Show fit
|
0.10 |
ENST00000394013.6
ENST00000345093.9
ENST00000551711.5
ENST00000552656.5
|
RNF41
|
ring finger protein 41
|
|
chr11_-_114400417
Show fit
|
0.10 |
ENST00000325636.8
ENST00000623205.2
|
C11orf71
|
chromosome 11 open reading frame 71
|
|
chr11_+_8911154
Show fit
|
0.10 |
ENST00000309357.8
ENST00000529876.5
ENST00000309377.9
ENST00000525005.5
ENST00000524577.5
ENST00000534506.5
|
AKIP1
|
A-kinase interacting protein 1
|
|
chr4_-_8440712
Show fit
|
0.09 |
ENST00000356406.10
ENST00000413009.6
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr11_+_8911106
Show fit
|
0.09 |
ENST00000299576.9
|
AKIP1
|
A-kinase interacting protein 1
|
|
chr19_-_42412347
Show fit
|
0.09 |
ENST00000601189.1
ENST00000599211.1
|
LIPE
|
lipase E, hormone sensitive type
|
|
chr11_+_8911280
Show fit
|
0.09 |
ENST00000530281.5
ENST00000396648.6
ENST00000534147.5
ENST00000529942.1
|
AKIP1
|
A-kinase interacting protein 1
|
|
chr6_-_26234978
Show fit
|
0.08 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member
|
|
chr22_+_21567705
Show fit
|
0.08 |
ENST00000342192.9
|
UBE2L3
|
ubiquitin conjugating enzyme E2 L3
|
|
chr19_+_55676621
Show fit
|
0.08 |
ENST00000411543.6
|
EPN1
|
epsin 1
|
|
chr19_-_14835162
Show fit
|
0.08 |
ENST00000322301.5
|
OR7A5
|
olfactory receptor family 7 subfamily A member 5
|
|
chrX_-_8171267
Show fit
|
0.08 |
ENST00000317103.5
|
VCX2
|
variable charge X-linked 2
|
|
chr3_-_119677346
Show fit
|
0.07 |
ENST00000484810.5
ENST00000497116.1
ENST00000261070.7
|
COX17
|
cytochrome c oxidase copper chaperone COX17
|
|
chr20_-_37158904
Show fit
|
0.07 |
ENST00000417458.5
|
MROH8
|
maestro heat like repeat family member 8
|
|
chrX_-_18220878
Show fit
|
0.07 |
ENST00000380033.9
|
BEND2
|
BEN domain containing 2
|
|
chr17_-_16569169
Show fit
|
0.07 |
ENST00000395824.5
|
ZNF287
|
zinc finger protein 287
|
|
chr14_-_106411021
Show fit
|
0.07 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional)
|
|
chr18_-_50819764
Show fit
|
0.06 |
ENST00000592966.5
|
MRO
|
maestro
|
|
chr5_-_173616588
Show fit
|
0.06 |
ENST00000285908.5
ENST00000311086.9
ENST00000480951.1
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chr4_-_75672868
Show fit
|
0.06 |
ENST00000678123.1
ENST00000678578.1
ENST00000677876.1
ENST00000676839.1
ENST00000678265.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr17_-_16569184
Show fit
|
0.06 |
ENST00000448349.2
ENST00000395825.4
|
ZNF287
|
zinc finger protein 287
|
|
chr4_+_37891060
Show fit
|
0.06 |
ENST00000261439.9
ENST00000508802.5
ENST00000402522.1
|
TBC1D1
|
TBC1 domain family member 1
|
|
chr10_-_27983103
Show fit
|
0.06 |
ENST00000672841.1
|
ODAD2
|
outer dynein arm docking complex subunit 2
|
|
chr11_-_75669028
Show fit
|
0.06 |
ENST00000304771.8
|
MAP6
|
microtubule associated protein 6
|
|
chr14_-_106538331
Show fit
|
0.06 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48
|
|
chr6_+_18387326
Show fit
|
0.05 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B
|
|
chr17_-_63918817
Show fit
|
0.05 |
ENST00000458650.6
ENST00000351388.8
ENST00000342364.8
ENST00000617086.1
ENST00000323322.10
ENST00000392824.8
|
GH1
CSHL1
|
growth hormone 1
chorionic somatomammotropin hormone like 1
|
|
chr18_+_58862904
Show fit
|
0.05 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532
|
|
chr4_-_185425941
Show fit
|
0.05 |
ENST00000264689.11
ENST00000505357.1
|
UFSP2
|
UFM1 specific peptidase 2
|
|
chr16_-_4802615
Show fit
|
0.05 |
ENST00000591392.5
ENST00000587711.5
ENST00000322048.12
|
ROGDI
|
rogdi atypical leucine zipper
|
|
chr17_+_43171187
Show fit
|
0.05 |
ENST00000542611.5
ENST00000590996.6
ENST00000589872.1
|
NBR1
|
NBR1 autophagy cargo receptor
|
|
chr1_+_145719483
Show fit
|
0.05 |
ENST00000369288.7
|
CD160
|
CD160 molecule
|
|
chr9_-_35563867
Show fit
|
0.05 |
ENST00000399742.7
ENST00000619051.4
|
FAM166B
|
family with sequence similarity 166 member B
|
|
chr2_-_37324826
Show fit
|
0.05 |
ENST00000234179.8
|
PRKD3
|
protein kinase D3
|
|
chr18_+_7754959
Show fit
|
0.04 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M
|
|
chr16_+_8642375
Show fit
|
0.04 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22
|
|
chr11_-_506739
Show fit
|
0.04 |
ENST00000529306.5
ENST00000438658.6
ENST00000527485.5
ENST00000397615.6
ENST00000397614.5
|
RNH1
|
ribonuclease/angiogenin inhibitor 1
|
|
chr2_+_241637682
Show fit
|
0.04 |
ENST00000419606.5
ENST00000404914.8
ENST00000400771.7
|
ATG4B
|
autophagy related 4B cysteine peptidase
|
|
chr1_+_1915080
Show fit
|
0.04 |
ENST00000378604.3
|
CALML6
|
calmodulin like 6
|
|
chr1_+_75796867
Show fit
|
0.04 |
ENST00000263187.4
|
MSH4
|
mutS homolog 4
|
|
chr8_+_24440930
Show fit
|
0.04 |
ENST00000441335.6
ENST00000175238.10
ENST00000380789.5
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr15_-_19988117
Show fit
|
0.03 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene)
|
|
chr19_+_58059236
Show fit
|
0.03 |
ENST00000359978.10
ENST00000401053.8
ENST00000511556.5
ENST00000506786.1
ENST00000313434.10
|
ZNF135
|
zinc finger protein 135
|
|
chr1_+_93345893
Show fit
|
0.03 |
ENST00000370272.9
ENST00000370267.1
|
DR1
|
down-regulator of transcription 1
|
|
chr17_+_20579724
Show fit
|
0.03 |
ENST00000661883.1
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15 like 2
|
|
chr11_+_114400592
Show fit
|
0.03 |
ENST00000541475.5
|
RBM7
|
RNA binding motif protein 7
|
|
chr2_-_230960954
Show fit
|
0.03 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55
|
|
chr11_+_114400646
Show fit
|
0.02 |
ENST00000375490.10
ENST00000544582.1
ENST00000545678.1
|
RBM7
|
RNA binding motif protein 7
|
|
chr16_+_56638659
Show fit
|
0.02 |
ENST00000290705.12
|
MT1A
|
metallothionein 1A
|
|
chrX_+_8465426
Show fit
|
0.02 |
ENST00000381029.4
|
VCX3B
|
variable charge X-linked 3B
|
|
chr12_-_10098977
Show fit
|
0.02 |
ENST00000315330.8
ENST00000457018.6
|
CLEC1A
|
C-type lectin domain family 1 member A
|
|
chr14_-_45253402
Show fit
|
0.02 |
ENST00000627697.1
|
MIS18BP1
|
MIS18 binding protein 1
|
|
chr14_+_73886617
Show fit
|
0.02 |
ENST00000540593.5
ENST00000555730.6
|
ZNF410
|
zinc finger protein 410
|
|
chr2_-_108989206
Show fit
|
0.02 |
ENST00000258443.7
ENST00000409271.5
ENST00000376651.1
|
EDAR
|
ectodysplasin A receptor
|
|
chr16_+_56608577
Show fit
|
0.02 |
ENST00000245185.6
ENST00000561491.1
|
MT2A
|
metallothionein 2A
|
|
chr14_-_106658251
Show fit
|
0.02 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64
|
|
chr19_+_8052752
Show fit
|
0.01 |
ENST00000315626.6
ENST00000253451.9
|
CCL25
|
C-C motif chemokine ligand 25
|
|
chr6_+_127577168
Show fit
|
0.01 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr11_+_114400030
Show fit
|
0.01 |
ENST00000540163.5
|
RBM7
|
RNA binding motif protein 7
|
|
chr17_-_63873676
Show fit
|
0.01 |
ENST00000613718.3
ENST00000392886.7
ENST00000345366.8
ENST00000336844.9
ENST00000560142.5
|
CSH2
|
chorionic somatomammotropin hormone 2
|
|
chr1_-_48776811
Show fit
|
0.01 |
ENST00000371833.4
|
BEND5
|
BEN domain containing 5
|
|
chrX_+_2691270
Show fit
|
0.01 |
ENST00000381187.8
ENST00000381184.6
|
CD99
|
CD99 molecule (Xg blood group)
|
|
chr19_+_8053000
Show fit
|
0.01 |
ENST00000390669.7
|
CCL25
|
C-C motif chemokine ligand 25
|
|
chr3_+_13480215
Show fit
|
0.01 |
ENST00000446613.6
ENST00000402259.5
ENST00000402271.5
ENST00000295757.8
ENST00000404548.5
ENST00000404040.5
|
HDAC11
|
histone deacetylase 11
|
|
chr16_-_67944113
Show fit
|
0.01 |
ENST00000264005.10
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr2_+_190180930
Show fit
|
0.01 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chrX_-_24027186
Show fit
|
0.01 |
ENST00000328046.8
|
KLHL15
|
kelch like family member 15
|
|
chr22_+_23688133
Show fit
|
0.01 |
ENST00000401461.5
|
RGL4
|
ral guanine nucleotide dissociation stimulator like 4
|
|
chr8_-_48921419
Show fit
|
0.01 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2
|
|
chrX_+_2691310
Show fit
|
0.01 |
ENST00000482405.7
ENST00000624481.4
ENST00000381180.9
|
CD99
|
CD99 molecule (Xg blood group)
|
|
chr16_+_32066065
Show fit
|
0.01 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional)
|
|
chr17_-_63896621
Show fit
|
0.00 |
ENST00000453363.7
|
CSH1
|
chorionic somatomammotropin hormone 1
|
|
chrX_+_2691164
Show fit
|
0.00 |
ENST00000611428.5
|
CD99
|
CD99 molecule (Xg blood group)
|
|
chr4_+_87006988
Show fit
|
0.00 |
ENST00000307808.10
|
AFF1
|
AF4/FMR2 family member 1
|
|
chr8_-_22156789
Show fit
|
0.00 |
ENST00000306317.7
|
LGI3
|
leucine rich repeat LGI family member 3
|
|
chrX_+_2691284
Show fit
|
0.00 |
ENST00000381192.10
|
CD99
|
CD99 molecule (Xg blood group)
|
|
chr2_-_74648307
Show fit
|
0.00 |
ENST00000421985.2
ENST00000536235.5
|
M1AP
|
meiosis 1 associated protein
|