Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
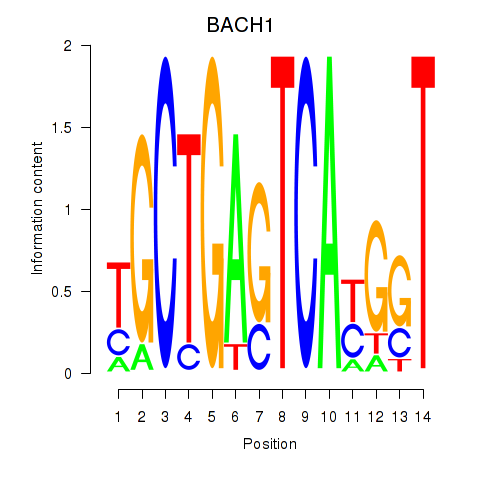
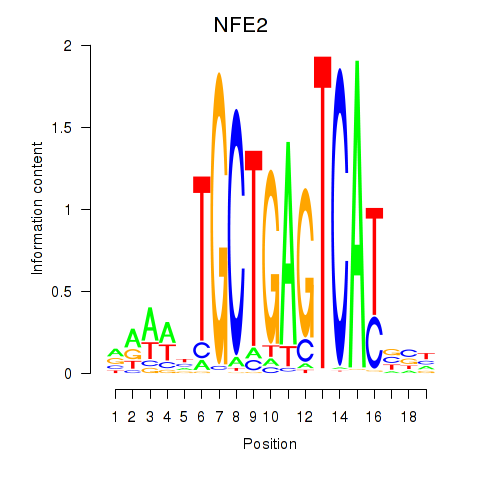
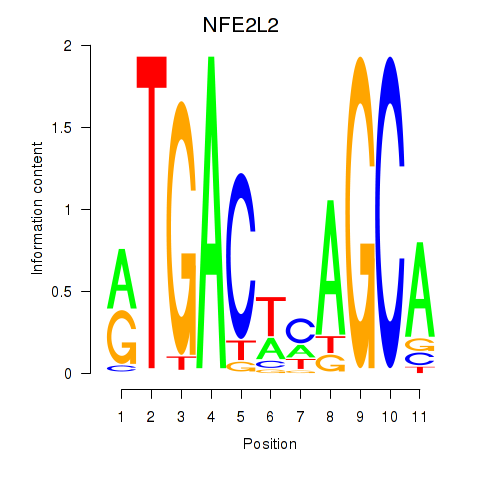
Results for BACH1_NFE2_NFE2L2
Z-value: 0.27
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.16 | BTB domain and CNC homolog 1 |
|
NFE2
|
ENSG00000123405.14 | nuclear factor, erythroid 2 |
|
NFE2L2
|
ENSG00000116044.16 | nuclear factor, erythroid 2 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BACH1 | hg38_v1_chr21_+_29299368_29299441, hg38_v1_chr21_+_29298890_29298932 | 0.61 | 1.1e-01 | Click! |
| NFE2L2 | hg38_v1_chr2_-_177263522_177263572, hg38_v1_chr2_-_177263800_177263870 | -0.40 | 3.3e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_55681647 | 0.68 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B |
| chr12_+_55681711 | 0.67 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr10_-_125816510 | 0.62 |
ENST00000650587.1
|
UROS
|
uroporphyrinogen III synthase |
| chr10_-_125816596 | 0.58 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr8_-_30727777 | 0.47 |
ENST00000537535.5
ENST00000541648.5 ENST00000546342.5 ENST00000221130.11 |
GSR
|
glutathione-disulfide reductase |
| chr11_-_2903490 | 0.45 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chr5_-_150155828 | 0.40 |
ENST00000261799.9
|
PDGFRB
|
platelet derived growth factor receptor beta |
| chr10_-_75109085 | 0.40 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr14_+_21990357 | 0.38 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr5_+_179820895 | 0.38 |
ENST00000504627.1
ENST00000389805.9 ENST00000510187.5 |
SQSTM1
|
sequestosome 1 |
| chr12_+_10213417 | 0.37 |
ENST00000546017.5
ENST00000535576.5 ENST00000539170.5 |
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr10_-_75109172 | 0.36 |
ENST00000372700.7
ENST00000473072.2 ENST00000491677.6 ENST00000372702.7 |
DUSP13
|
dual specificity phosphatase 13 |
| chr2_+_54115437 | 0.35 |
ENST00000303536.8
ENST00000394666.7 |
ACYP2
|
acylphosphatase 2 |
| chr19_-_58511981 | 0.30 |
ENST00000263093.7
ENST00000601355.1 |
SLC27A5
|
solute carrier family 27 member 5 |
| chr16_-_69726506 | 0.29 |
ENST00000561500.5
ENST00000320623.10 ENST00000439109.6 ENST00000564043.1 ENST00000379046.6 ENST00000379047.7 |
NQO1
|
NAD(P)H quinone dehydrogenase 1 |
| chr7_+_134527560 | 0.29 |
ENST00000359579.5
|
AKR1B10
|
aldo-keto reductase family 1 member B10 |
| chr2_+_54115396 | 0.28 |
ENST00000406041.5
|
ACYP2
|
acylphosphatase 2 |
| chrX_-_15493234 | 0.28 |
ENST00000380420.10
|
PIR
|
pirin |
| chr7_+_22727147 | 0.28 |
ENST00000426291.5
ENST00000401651.5 ENST00000258743.10 ENST00000407492.5 ENST00000401630.7 ENST00000406575.1 |
IL6
|
interleukin 6 |
| chr4_-_993376 | 0.27 |
ENST00000398520.6
ENST00000398516.3 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr19_+_18386150 | 0.26 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr1_-_45521931 | 0.25 |
ENST00000447184.6
ENST00000262746.5 |
PRDX1
|
peroxiredoxin 1 |
| chr1_-_45521854 | 0.25 |
ENST00000372079.1
ENST00000319248.13 |
PRDX1
|
peroxiredoxin 1 |
| chr6_-_35921079 | 0.25 |
ENST00000507909.1
ENST00000373825.7 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_-_35921128 | 0.24 |
ENST00000510290.5
ENST00000423325.6 |
SRPK1
|
SRSF protein kinase 1 |
| chr14_-_64942720 | 0.23 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chr16_+_2033264 | 0.23 |
ENST00000565855.5
ENST00000566198.1 |
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr19_+_18588789 | 0.23 |
ENST00000450195.6
ENST00000358607.11 |
REX1BD
|
required for excision 1-B domain containing |
| chr17_-_75154534 | 0.22 |
ENST00000356033.8
|
JPT1
|
Jupiter microtubule associated homolog 1 |
| chr16_+_30065753 | 0.22 |
ENST00000642816.3
ENST00000643777.4 ENST00000569798.5 |
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr2_-_210315160 | 0.21 |
ENST00000352451.4
|
MYL1
|
myosin light chain 1 |
| chr17_+_18183052 | 0.20 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr21_+_31659666 | 0.20 |
ENST00000389995.4
ENST00000270142.11 |
SOD1
|
superoxide dismutase 1 |
| chr14_-_64942783 | 0.20 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr1_-_243843226 | 0.20 |
ENST00000336199.9
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr4_-_993430 | 0.20 |
ENST00000361661.6
ENST00000622731.4 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr17_-_35063648 | 0.20 |
ENST00000394597.7
|
RFFL
|
ring finger and FYVE like domain containing E3 ubiquitin protein ligase |
| chr11_-_111923722 | 0.20 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr10_-_91633057 | 0.19 |
ENST00000238994.6
|
PPP1R3C
|
protein phosphatase 1 regulatory subunit 3C |
| chr19_+_46303599 | 0.19 |
ENST00000300862.7
|
HIF3A
|
hypoxia inducible factor 3 subunit alpha |
| chr16_+_30065777 | 0.18 |
ENST00000395240.7
ENST00000566846.5 |
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr5_-_177496802 | 0.18 |
ENST00000506161.5
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr10_-_5977492 | 0.18 |
ENST00000530685.5
ENST00000397255.7 ENST00000379971.5 ENST00000528354.5 ENST00000397250.6 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr8_-_100336184 | 0.18 |
ENST00000519527.5
ENST00000522369.5 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr17_+_81977587 | 0.17 |
ENST00000306739.9
ENST00000581647.5 ENST00000580534.5 ENST00000579684.5 |
ASPSCR1
|
ASPSCR1 tether for SLC2A4, UBX domain containing |
| chr5_-_177496845 | 0.17 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr17_-_30291930 | 0.17 |
ENST00000261714.11
|
BLMH
|
bleomycin hydrolase |
| chr12_+_12357056 | 0.16 |
ENST00000314565.9
ENST00000542728.5 |
BORCS5
|
BLOC-1 related complex subunit 5 |
| chr6_-_35921047 | 0.16 |
ENST00000361690.7
ENST00000512445.5 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_-_132659178 | 0.16 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr5_-_142685654 | 0.16 |
ENST00000378046.5
ENST00000619447.4 |
FGF1
|
fibroblast growth factor 1 |
| chr11_-_47185840 | 0.16 |
ENST00000539589.5
ENST00000528462.5 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_-_102955705 | 0.16 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr22_+_18906303 | 0.16 |
ENST00000331444.12
ENST00000608842.1 |
DGCR6
|
DiGeorge syndrome critical region gene 6 |
| chr20_-_653189 | 0.15 |
ENST00000381962.4
|
SRXN1
|
sulfiredoxin 1 |
| chr16_+_89921851 | 0.15 |
ENST00000554444.5
ENST00000556565.5 |
TUBB3
|
tubulin beta 3 class III |
| chr11_-_707063 | 0.15 |
ENST00000683307.1
|
DEAF1
|
DEAF1 transcription factor |
| chr6_-_28354193 | 0.15 |
ENST00000396838.6
ENST00000426434.1 ENST00000434036.5 ENST00000439628.5 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr8_-_132111159 | 0.15 |
ENST00000673615.1
ENST00000434736.6 |
HHLA1
|
HERV-H LTR-associating 1 |
| chrX_+_154542194 | 0.15 |
ENST00000618670.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr14_+_58244821 | 0.14 |
ENST00000216455.9
ENST00000412908.6 ENST00000557508.5 |
PSMA3
|
proteasome 20S subunit alpha 3 |
| chr11_-_9003994 | 0.14 |
ENST00000309166.8
ENST00000525100.1 ENST00000531090.5 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr10_+_35195843 | 0.14 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr5_+_55102635 | 0.14 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chrX_+_12975083 | 0.14 |
ENST00000451311.7
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4 X-linked |
| chr9_+_100429511 | 0.14 |
ENST00000613183.1
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr3_-_48016400 | 0.14 |
ENST00000434267.5
ENST00000683076.1 ENST00000633710.1 |
MAP4
|
microtubule associated protein 4 |
| chr10_+_84139491 | 0.13 |
ENST00000372134.6
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr18_+_11857440 | 0.13 |
ENST00000602628.1
|
GNAL
|
G protein subunit alpha L |
| chr9_-_137028223 | 0.13 |
ENST00000341511.11
|
ABCA2
|
ATP binding cassette subfamily A member 2 |
| chr19_-_45782388 | 0.13 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr3_+_69866217 | 0.13 |
ENST00000314589.10
|
MITF
|
melanocyte inducing transcription factor |
| chr8_+_106447918 | 0.13 |
ENST00000442977.6
|
OXR1
|
oxidation resistance 1 |
| chr3_-_150022799 | 0.13 |
ENST00000649949.1
ENST00000494827.5 |
PFN2
|
profilin 2 |
| chr9_+_137188646 | 0.12 |
ENST00000322310.10
|
SSNA1
|
SS nuclear autoantigen 1 |
| chr8_-_26867267 | 0.12 |
ENST00000380573.4
|
ADRA1A
|
adrenoceptor alpha 1A |
| chr3_-_149576203 | 0.12 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr9_+_137188697 | 0.12 |
ENST00000464553.2
|
SSNA1
|
SS nuclear autoantigen 1 |
| chr17_-_78925376 | 0.12 |
ENST00000262768.11
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chrX_+_22136552 | 0.12 |
ENST00000682888.1
ENST00000684356.1 |
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr1_+_52142044 | 0.12 |
ENST00000287727.8
ENST00000371591.2 |
ZFYVE9
|
zinc finger FYVE-type containing 9 |
| chr9_-_137028271 | 0.12 |
ENST00000265662.9
ENST00000371605.7 |
ABCA2
|
ATP binding cassette subfamily A member 2 |
| chr1_+_156786875 | 0.12 |
ENST00000526188.5
ENST00000454659.1 |
PRCC
|
proline rich mitotic checkpoint control factor |
| chr9_+_72577788 | 0.12 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr1_-_51331315 | 0.12 |
ENST00000262676.9
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr3_+_51943244 | 0.12 |
ENST00000498510.2
|
PARP3
|
poly(ADP-ribose) polymerase family member 3 |
| chr17_-_39730496 | 0.11 |
ENST00000577810.1
|
MIEN1
|
migration and invasion enhancer 1 |
| chr10_-_70132801 | 0.11 |
ENST00000307864.3
ENST00000613322.4 |
AIFM2
|
apoptosis inducing factor mitochondria associated 2 |
| chr9_-_70414657 | 0.11 |
ENST00000377126.4
|
KLF9
|
Kruppel like factor 9 |
| chr17_-_39730524 | 0.11 |
ENST00000394231.8
|
MIEN1
|
migration and invasion enhancer 1 |
| chr17_+_45221993 | 0.11 |
ENST00000328118.7
|
FMNL1
|
formin like 1 |
| chr3_+_46163465 | 0.11 |
ENST00000357422.2
|
CCR3
|
C-C motif chemokine receptor 3 |
| chr17_-_15566276 | 0.11 |
ENST00000630868.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr2_+_234050732 | 0.11 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2 |
| chr2_-_96505345 | 0.11 |
ENST00000310865.7
ENST00000451794.6 |
NEURL3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr9_+_2158487 | 0.11 |
ENST00000634706.1
ENST00000634338.1 ENST00000635688.1 ENST00000634435.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_-_23034878 | 0.10 |
ENST00000493471.2
ENST00000460922.2 ENST00000361611.11 |
PSMB5
|
proteasome 20S subunit beta 5 |
| chr6_+_63521738 | 0.10 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr19_-_45639104 | 0.10 |
ENST00000586770.5
ENST00000591721.5 ENST00000245925.8 ENST00000590043.5 ENST00000589876.5 |
EML2
|
EMAP like 2 |
| chr12_-_120250145 | 0.10 |
ENST00000458477.6
|
PXN
|
paxillin |
| chrX_+_150363258 | 0.10 |
ENST00000683696.1
|
MAMLD1
|
mastermind like domain containing 1 |
| chrX_+_150363306 | 0.10 |
ENST00000370401.7
ENST00000432680.7 |
MAMLD1
|
mastermind like domain containing 1 |
| chr10_-_25062279 | 0.10 |
ENST00000615958.4
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr9_+_72577369 | 0.10 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr11_+_119087979 | 0.10 |
ENST00000535253.5
ENST00000392841.1 ENST00000648374.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr1_-_243843164 | 0.10 |
ENST00000491219.6
ENST00000680056.1 ENST00000492957.2 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr14_-_75069478 | 0.09 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr22_+_22697789 | 0.09 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr17_+_77376083 | 0.09 |
ENST00000427674.6
|
SEPTIN9
|
septin 9 |
| chr2_-_42764116 | 0.09 |
ENST00000378661.3
|
OXER1
|
oxoeicosanoid receptor 1 |
| chr2_-_191020960 | 0.09 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1 |
| chr17_+_18183803 | 0.09 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr22_+_44676808 | 0.09 |
ENST00000624862.3
|
PRR5
|
proline rich 5 |
| chr4_+_17614630 | 0.09 |
ENST00000237380.12
|
MED28
|
mediator complex subunit 28 |
| chr15_-_82647960 | 0.09 |
ENST00000615198.4
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr9_-_127874964 | 0.08 |
ENST00000373156.5
|
AK1
|
adenylate kinase 1 |
| chr16_-_1379674 | 0.08 |
ENST00000402641.6
ENST00000248104.11 ENST00000674376.1 ENST00000397464.5 |
UNKL
|
unk like zinc finger |
| chr17_+_21288029 | 0.08 |
ENST00000526076.6
ENST00000361818.9 ENST00000316920.10 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr21_+_32298849 | 0.08 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_-_71725365 | 0.08 |
ENST00000425534.8
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr6_+_56955097 | 0.08 |
ENST00000370746.8
ENST00000370748.7 |
BEND6
|
BEN domain containing 6 |
| chr16_+_11965193 | 0.08 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr5_+_150640652 | 0.08 |
ENST00000307662.5
|
SYNPO
|
synaptopodin |
| chr19_-_38899529 | 0.08 |
ENST00000249396.12
ENST00000437828.5 |
SIRT2
|
sirtuin 2 |
| chr22_-_45212431 | 0.08 |
ENST00000496226.1
ENST00000251993.11 |
KIAA0930
|
KIAA0930 |
| chr17_+_7440738 | 0.08 |
ENST00000575398.5
ENST00000575082.5 |
FGF11
|
fibroblast growth factor 11 |
| chr10_+_68106109 | 0.08 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr10_+_132066003 | 0.08 |
ENST00000657318.1
ENST00000666210.1 |
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr10_-_70146602 | 0.08 |
ENST00000335494.5
ENST00000287078.7 |
TYSND1
|
trypsin like peroxisomal matrix peptidase 1 |
| chr14_+_79279403 | 0.08 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr3_+_126704202 | 0.08 |
ENST00000290913.8
ENST00000508789.5 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr1_-_162868761 | 0.08 |
ENST00000367910.5
ENST00000367912.6 |
CCDC190
|
coiled-coil domain containing 190 |
| chr14_+_34993240 | 0.08 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr16_+_29808125 | 0.07 |
ENST00000568282.1
|
MAZ
|
MYC associated zinc finger protein |
| chr22_+_22704265 | 0.07 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr4_-_165279679 | 0.07 |
ENST00000505354.2
|
GK3P
|
glycerol kinase 3 pseudogene |
| chr10_-_50279715 | 0.07 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chr16_+_8642375 | 0.07 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr8_-_100105955 | 0.07 |
ENST00000523437.5
|
RGS22
|
regulator of G protein signaling 22 |
| chr19_-_38899800 | 0.07 |
ENST00000414941.5
ENST00000358931.9 ENST00000392081.6 |
SIRT2
|
sirtuin 2 |
| chr2_+_89851723 | 0.07 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr20_+_17699942 | 0.07 |
ENST00000427254.1
ENST00000377805.7 |
BANF2
|
BANF family member 2 |
| chr1_-_46132650 | 0.07 |
ENST00000372006.5
ENST00000425892.2 ENST00000420542.5 |
PIK3R3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr1_-_150235972 | 0.07 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr12_+_104286881 | 0.07 |
ENST00000526691.5
ENST00000531691.5 ENST00000526390.5 ENST00000531689.5 |
TXNRD1
|
thioredoxin reductase 1 |
| chr3_+_191329342 | 0.07 |
ENST00000392455.9
|
CCDC50
|
coiled-coil domain containing 50 |
| chr1_-_235588959 | 0.07 |
ENST00000484517.2
|
GNG4
|
G protein subunit gamma 4 |
| chr22_-_30667795 | 0.07 |
ENST00000404885.5
ENST00000403268.1 ENST00000407308.1 ENST00000334679.4 ENST00000342474.4 |
DUSP18
|
dual specificity phosphatase 18 |
| chr6_-_53510445 | 0.07 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chr10_+_132065937 | 0.07 |
ENST00000658847.1
ENST00000666974.1 |
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr16_+_74296789 | 0.07 |
ENST00000219313.9
ENST00000567958.5 ENST00000568615.2 |
PSMD7
|
proteasome 26S subunit, non-ATPase 7 |
| chr11_+_93741591 | 0.07 |
ENST00000528288.5
ENST00000617482.4 ENST00000540113.5 |
C11orf54
|
chromosome 11 open reading frame 54 |
| chr4_+_15002443 | 0.06 |
ENST00000538197.7
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr2_-_219308963 | 0.06 |
ENST00000423636.6
ENST00000442029.5 ENST00000412847.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr1_-_15809348 | 0.06 |
ENST00000483273.2
|
UQCRHL
|
ubiquinol-cytochrome c reductase hinge protein like |
| chr2_-_169031317 | 0.06 |
ENST00000650372.1
|
ABCB11
|
ATP binding cassette subfamily B member 11 |
| chr3_+_184186023 | 0.06 |
ENST00000429586.6
ENST00000292808.5 |
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr9_+_2158239 | 0.06 |
ENST00000635133.1
ENST00000634931.1 ENST00000423555.6 ENST00000382185.6 ENST00000302401.8 ENST00000382183.6 ENST00000417599.6 ENST00000382186.6 ENST00000635530.1 ENST00000635388.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrX_-_24647091 | 0.06 |
ENST00000356768.8
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr6_+_86937500 | 0.06 |
ENST00000305344.7
|
HTR1E
|
5-hydroxytryptamine receptor 1E |
| chr11_+_10456186 | 0.06 |
ENST00000528723.5
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr5_+_150640678 | 0.06 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr9_-_114078293 | 0.06 |
ENST00000265132.8
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr11_+_20363780 | 0.06 |
ENST00000532505.1
|
HTATIP2
|
HIV-1 Tat interactive protein 2 |
| chr2_+_90172802 | 0.06 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr16_+_87392327 | 0.06 |
ENST00000565788.1
ENST00000268607.10 |
MAP1LC3B
|
microtubule associated protein 1 light chain 3 beta |
| chr19_+_38374758 | 0.06 |
ENST00000585598.1
ENST00000602911.5 ENST00000592561.5 |
PSMD8
|
proteasome 26S subunit, non-ATPase 8 |
| chr10_-_72217126 | 0.06 |
ENST00000317126.8
|
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr2_-_89027700 | 0.06 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr20_-_1329131 | 0.06 |
ENST00000360779.4
|
SDCBP2
|
syndecan binding protein 2 |
| chr17_+_4950147 | 0.06 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr3_-_170870033 | 0.06 |
ENST00000466674.5
ENST00000295830.13 ENST00000463836.1 |
RPL22L1
|
ribosomal protein L22 like 1 |
| chr19_+_38374557 | 0.06 |
ENST00000215071.9
|
PSMD8
|
proteasome 26S subunit, non-ATPase 8 |
| chr9_+_2157647 | 0.06 |
ENST00000452193.5
ENST00000324954.10 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_-_110256466 | 0.06 |
ENST00000374515.9
ENST00000374517.6 |
TXN
|
thioredoxin |
| chr1_-_17045219 | 0.06 |
ENST00000491274.5
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr5_+_161848112 | 0.06 |
ENST00000393943.10
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr4_-_86453278 | 0.06 |
ENST00000638867.2
ENST00000642019.1 ENST00000641313.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_-_100428657 | 0.06 |
ENST00000360951.8
ENST00000398027.6 ENST00000684423.1 ENST00000472716.1 |
ZCWPW1
|
zinc finger CW-type and PWWP domain containing 1 |
| chr1_-_150235943 | 0.05 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chrX_+_136487940 | 0.05 |
ENST00000370648.4
|
BRS3
|
bombesin receptor subtype 3 |
| chr22_+_44677077 | 0.05 |
ENST00000403581.5
|
PRR5
|
proline rich 5 |
| chr19_+_38374549 | 0.05 |
ENST00000620216.4
|
PSMD8
|
proteasome 26S subunit, non-ATPase 8 |
| chr12_+_8157034 | 0.05 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr16_+_31117656 | 0.05 |
ENST00000219797.9
ENST00000448516.6 |
KAT8
|
lysine acetyltransferase 8 |
| chr6_+_44247087 | 0.05 |
ENST00000353801.7
ENST00000371646.10 |
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr18_-_57586668 | 0.05 |
ENST00000592699.6
ENST00000382873.8 ENST00000262093.11 ENST00000652755.1 |
FECH
|
ferrochelatase |
| chr1_+_44775531 | 0.05 |
ENST00000396651.8
ENST00000372209.3 |
RPS8
|
ribosomal protein S8 |
| chr13_+_108269629 | 0.05 |
ENST00000430559.5
ENST00000375887.9 |
TNFSF13B
|
TNF superfamily member 13b |
| chr19_+_38899680 | 0.05 |
ENST00000576510.5
ENST00000392079.7 |
NFKBIB
|
NFKB inhibitor beta |
| chr10_-_97292625 | 0.05 |
ENST00000466484.1
ENST00000358531.9 ENST00000358308.7 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr17_-_7219813 | 0.05 |
ENST00000399510.8
ENST00000648172.8 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr11_+_67583803 | 0.05 |
ENST00000398606.10
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr20_-_44210697 | 0.05 |
ENST00000255174.3
|
OSER1
|
oxidative stress responsive serine rich 1 |
| chr10_+_104254163 | 0.05 |
ENST00000539281.5
|
GSTO1
|
glutathione S-transferase omega 1 |
| chr6_-_83431038 | 0.05 |
ENST00000369705.4
|
ME1
|
malic enzyme 1 |
| chr22_+_22395005 | 0.05 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr19_+_16185380 | 0.05 |
ENST00000589852.5
ENST00000263384.12 ENST00000588367.5 ENST00000587351.1 |
FAM32A
|
family with sequence similarity 32 member A |
| chrX_+_30653478 | 0.05 |
ENST00000378945.7
ENST00000378941.4 |
GK
|
glycerol kinase |
| chrX_-_71106728 | 0.05 |
ENST00000374251.6
|
CXorf65
|
chromosome X open reading frame 65 |
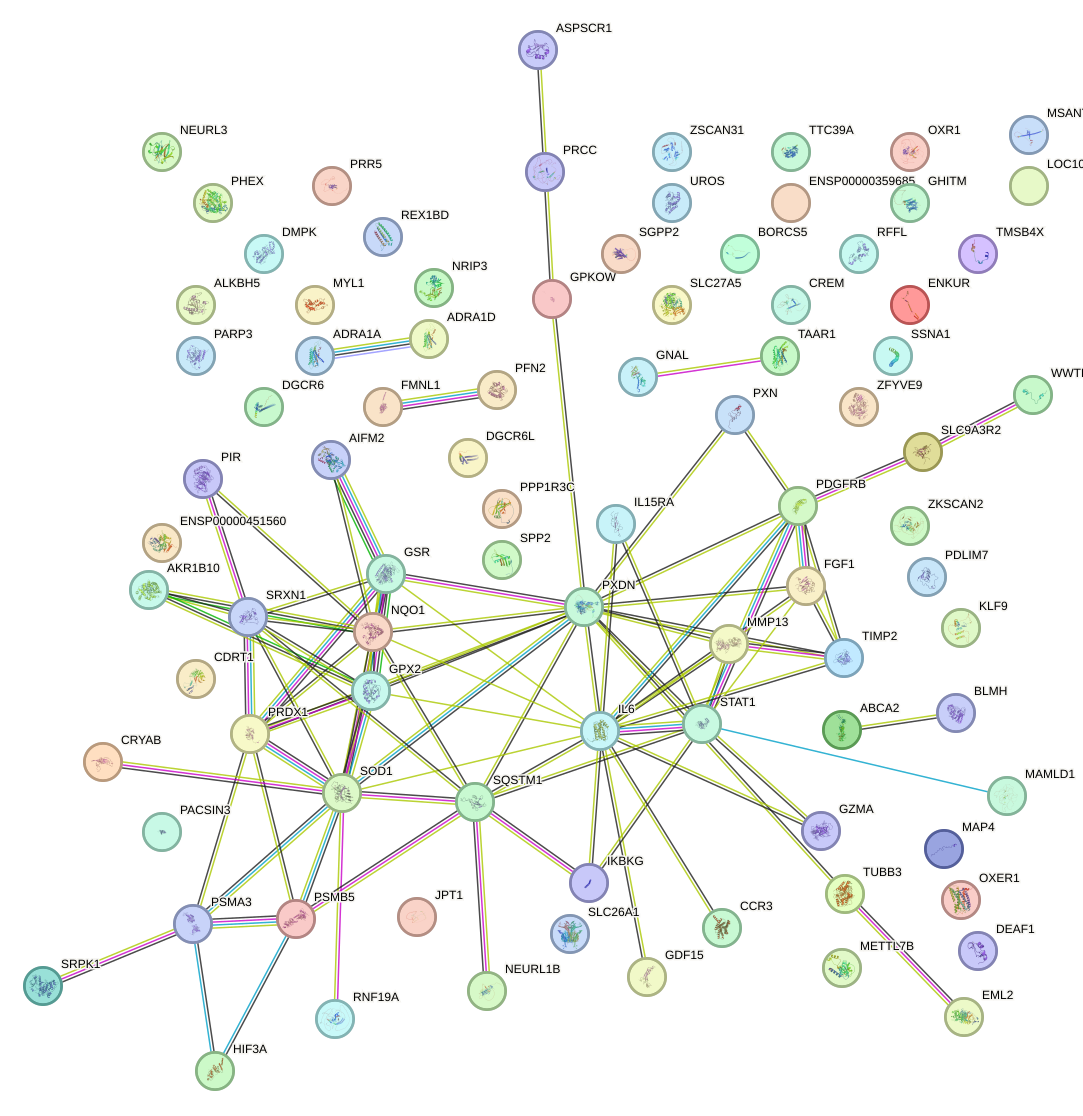
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.4 | GO:0072276 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 0.3 | GO:0016487 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.1 | 0.3 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.3 | GO:1990637 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 0.0 | 0.5 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.3 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.8 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.0 | 0.2 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 1.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.3 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.1 | 0.3 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.6 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |