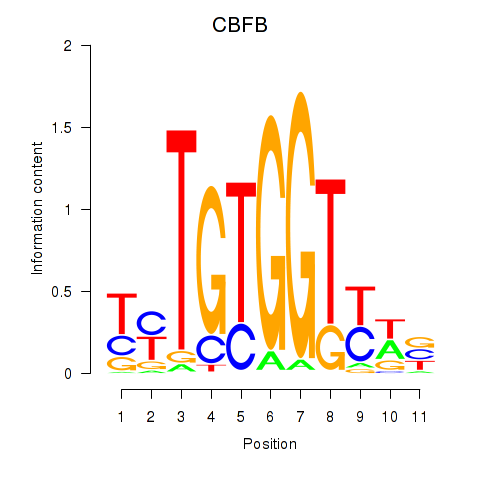
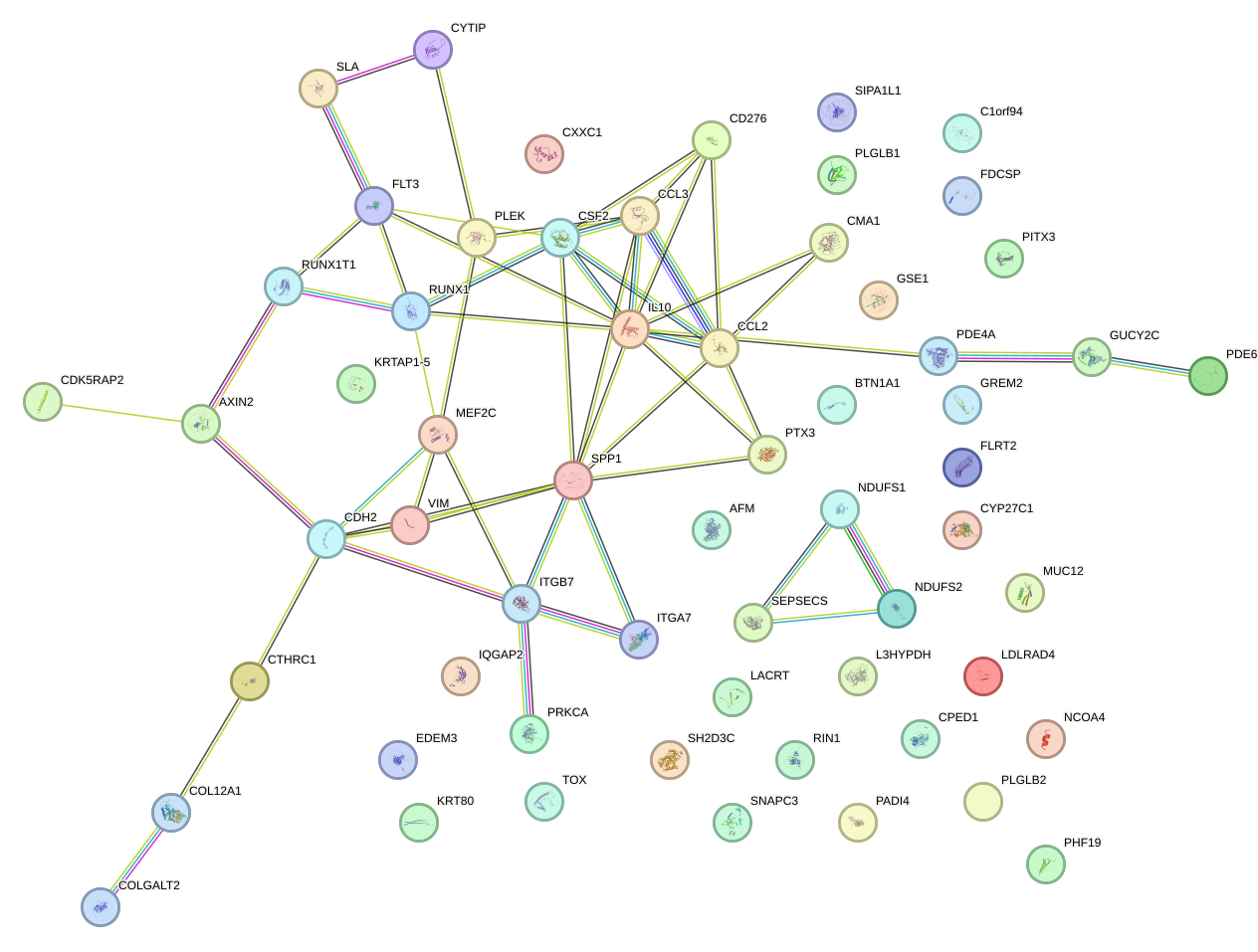
|
chr3_+_157436842
Show fit
|
0.33 |
ENST00000295927.4
|
PTX3
|
pentraxin 3
|
|
chr14_-_60649449
Show fit
|
0.30 |
ENST00000645694.3
|
SIX1
|
SIX homeobox 1
|
|
chr5_+_76403266
Show fit
|
0.22 |
ENST00000274364.11
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr8_+_103372388
Show fit
|
0.22 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr12_-_55707865
Show fit
|
0.20 |
ENST00000347027.10
ENST00000257879.11
ENST00000553804.6
|
ITGA7
|
integrin subunit alpha 7
|
|
chr8_-_59119121
Show fit
|
0.18 |
ENST00000361421.2
|
TOX
|
thymocyte selection associated high mobility group box
|
|
chr9_-_127778659
Show fit
|
0.18 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr13_+_75636311
Show fit
|
0.17 |
ENST00000377499.9
ENST00000377534.8
|
LMO7
|
LIM domain 7
|
|
chr2_-_157444044
Show fit
|
0.16 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr2_-_127220293
Show fit
|
0.16 |
ENST00000664447.2
ENST00000409327.2
|
CYP27C1
|
cytochrome P450 family 27 subfamily C member 1
|
|
chr18_-_28177934
Show fit
|
0.14 |
ENST00000676445.1
|
CDH2
|
cadherin 2
|
|
chr9_-_120580125
Show fit
|
0.13 |
ENST00000360190.8
ENST00000349780.9
ENST00000360822.7
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2
|
|
chr5_-_88785493
Show fit
|
0.12 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_75202792
Show fit
|
0.12 |
ENST00000416123.6
|
COL12A1
|
collagen type XII alpha 1 chain
|
|
chr6_+_26500296
Show fit
|
0.11 |
ENST00000684113.1
|
BTN1A1
|
butyrophilin subfamily 1 member A1
|
|
chr14_+_85530163
Show fit
|
0.10 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr22_+_22880706
Show fit
|
0.10 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1
|
|
chr14_+_85530127
Show fit
|
0.09 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr19_-_13938371
Show fit
|
0.08 |
ENST00000588872.3
ENST00000339560.10
|
PODNL1
|
podocan like 1
|
|
chr19_-_13937991
Show fit
|
0.08 |
ENST00000254320.7
ENST00000586075.1
|
PODNL1
|
podocan like 1
|
|
chr6_+_30940970
Show fit
|
0.08 |
ENST00000462446.6
ENST00000304311.3
|
MUCL3
|
mucin like 3
|
|
chr16_+_53054973
Show fit
|
0.07 |
ENST00000447540.6
ENST00000615216.4
ENST00000566029.5
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr1_+_17308194
Show fit
|
0.07 |
ENST00000375453.5
ENST00000375448.4
|
PADI4
|
peptidyl arginine deiminase 4
|
|
chr2_+_87748087
Show fit
|
0.07 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2
|
|
chr13_-_28100556
Show fit
|
0.07 |
ENST00000241453.12
|
FLT3
|
fms related receptor tyrosine kinase 3
|
|
chr14_-_24508251
Show fit
|
0.06 |
ENST00000250378.7
ENST00000206446.4
|
CMA1
|
chymase 1
|
|
chr17_+_34255274
Show fit
|
0.05 |
ENST00000580907.5
ENST00000225831.4
|
CCL2
|
C-C motif chemokine ligand 2
|
|
chr7_+_100969565
Show fit
|
0.05 |
ENST00000536621.6
ENST00000379442.7
|
MUC12
|
mucin 12, cell surface associated
|
|
chr10_+_17229267
Show fit
|
0.05 |
ENST00000224237.9
|
VIM
|
vimentin
|
|
chr17_+_66302606
Show fit
|
0.05 |
ENST00000413366.8
|
PRKCA
|
protein kinase C alpha
|
|
chr21_-_35049327
Show fit
|
0.04 |
ENST00000300305.7
|
RUNX1
|
RUNX family transcription factor 1
|
|
chr12_-_54634880
Show fit
|
0.04 |
ENST00000547511.5
ENST00000257867.5
|
LACRT
|
lacritin
|
|
chr7_+_120988683
Show fit
|
0.04 |
ENST00000340646.9
ENST00000310396.10
|
CPED1
|
cadherin like and PC-esterase domain containing 1
|
|
chr4_+_73481737
Show fit
|
0.04 |
ENST00000226355.5
|
AFM
|
afamin
|
|
chr12_-_14696571
Show fit
|
0.04 |
ENST00000261170.5
|
GUCY2C
|
guanylate cyclase 2C
|
|
chr1_-_184037695
Show fit
|
0.04 |
ENST00000361927.9
ENST00000649786.1
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2
|
|
chr9_-_120876356
Show fit
|
0.03 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19
|
|
chr12_-_52192007
Show fit
|
0.03 |
ENST00000394815.3
|
KRT80
|
keratin 80
|
|
chr21_-_35049238
Show fit
|
0.03 |
ENST00000416754.1
ENST00000437180.5
ENST00000455571.5
ENST00000675419.1
|
RUNX1
|
RUNX family transcription factor 1
|
|
chr2_-_206159410
Show fit
|
0.03 |
ENST00000457011.5
ENST00000440274.5
ENST00000432169.5
ENST00000233190.11
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1
|
|
chr12_-_52191981
Show fit
|
0.03 |
ENST00000313234.9
|
KRT80
|
keratin 80
|
|
chr17_-_36090133
Show fit
|
0.03 |
ENST00000613922.2
|
CCL3
|
C-C motif chemokine ligand 3
|
|
chr2_-_87021844
Show fit
|
0.03 |
ENST00000355705.4
ENST00000409310.6
|
PLGLB1
|
plasminogen like B1
|
|
chr1_+_34176900
Show fit
|
0.03 |
ENST00000488417.2
|
C1orf94
|
chromosome 1 open reading frame 94
|
|
chr17_-_41027198
Show fit
|
0.03 |
ENST00000361883.6
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr2_+_68365274
Show fit
|
0.02 |
ENST00000234313.8
|
PLEK
|
pleckstrin
|
|
chr12_-_53200443
Show fit
|
0.02 |
ENST00000550743.6
|
ITGB7
|
integrin subunit beta 7
|
|
chr10_-_46023445
Show fit
|
0.02 |
ENST00000585132.5
|
NCOA4
|
nuclear receptor coactivator 4
|
|
chr15_+_73683938
Show fit
|
0.02 |
ENST00000567189.5
|
CD276
|
CD276 molecule
|
|
chr18_+_13611910
Show fit
|
0.02 |
ENST00000590308.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr12_+_25052634
Show fit
|
0.02 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2
|
|
chr5_+_132073782
Show fit
|
0.02 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2
|
|
chr12_+_25052512
Show fit
|
0.01 |
ENST00000557489.5
ENST00000354454.7
ENST00000536173.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2
|
|
chr9_+_15422704
Show fit
|
0.01 |
ENST00000380821.7
ENST00000610884.4
ENST00000421710.5
|
SNAPC3
|
small nuclear RNA activating complex polypeptide 3
|
|
chr8_+_28890365
Show fit
|
0.01 |
ENST00000519662.5
ENST00000558662.5
ENST00000287701.15
ENST00000523613.5
ENST00000560599.5
ENST00000397358.7
|
HMBOX1
|
homeobox containing 1
|
|
chr16_+_85611401
Show fit
|
0.01 |
ENST00000405402.6
|
GSE1
|
Gse1 coiled-coil protein
|
|
chr4_+_70226116
Show fit
|
0.01 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein
|
|
chr2_-_206159194
Show fit
|
0.01 |
ENST00000455934.6
ENST00000449699.5
ENST00000454195.1
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1
|
|
chr1_-_206772484
Show fit
|
0.01 |
ENST00000423557.1
|
IL10
|
interleukin 10
|
|
chr11_-_66336396
Show fit
|
0.01 |
ENST00000627248.1
ENST00000311320.9
|
RIN1
|
Ras and Rab interactor 1
|
|
chr4_+_87975829
Show fit
|
0.01 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_-_240612147
Show fit
|
0.00 |
ENST00000318160.5
|
GREM2
|
gremlin 2, DAN family BMP antagonist
|
|
chr19_+_10430786
Show fit
|
0.00 |
ENST00000293683.9
|
PDE4A
|
phosphodiesterase 4A
|
|
chr17_-_65560296
Show fit
|
0.00 |
ENST00000585045.1
ENST00000611991.1
|
AXIN2
|
axin 2
|
|
chr1_-_184754808
Show fit
|
0.00 |
ENST00000318130.13
ENST00000367512.7
|
EDEM3
|
ER degradation enhancing alpha-mannosidase like protein 3
|
|
chr1_+_161197372
Show fit
|
0.00 |
ENST00000677550.1
ENST00000676600.1
ENST00000678507.1
ENST00000677579.1
ENST00000677231.1
ENST00000678911.1
ENST00000677846.1
|
NDUFS2
|
NADH:ubiquinone oxidoreductase core subunit S2
|
|
chr14_+_71598229
Show fit
|
0.00 |
ENST00000537413.5
|
SIPA1L1
|
signal induced proliferation associated 1 like 1
|
|
chr4_+_87975667
Show fit
|
0.00 |
ENST00000237623.11
ENST00000682655.1
ENST00000508233.6
ENST00000360804.4
ENST00000395080.8
|
SPP1
|
secreted phosphoprotein 1
|
|
chr14_-_59484317
Show fit
|
0.00 |
ENST00000247194.9
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase
|
|
chr8_-_133060347
Show fit
|
0.00 |
ENST00000427060.6
|
SLA
|
Src like adaptor
|
|
chr12_+_14973020
Show fit
|
0.00 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H
|