Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for DLX1_HOXA3_BARX2
Z-value: 0.94
Transcription factors associated with DLX1_HOXA3_BARX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
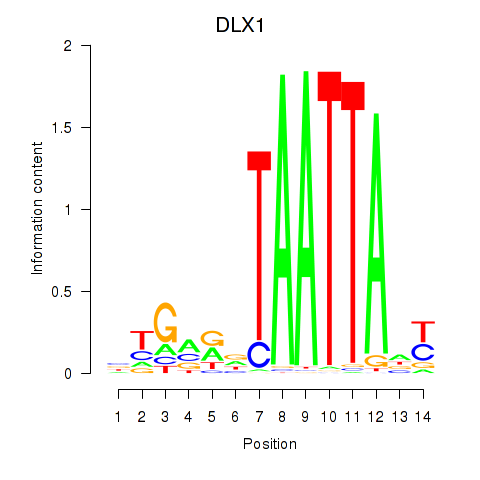
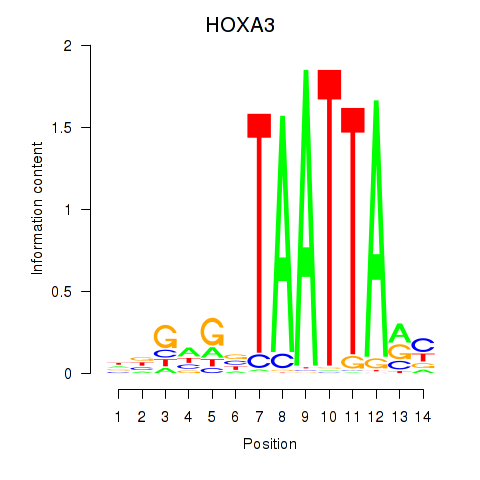
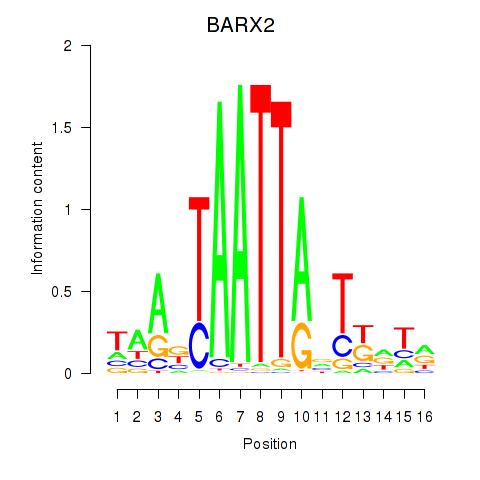
DLX1
|
ENSG00000144355.15 | distal-less homeobox 1 |
|
HOXA3
|
ENSG00000105997.23 | homeobox A3 |
|
BARX2
|
ENSG00000043039.7 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX2 | hg38_v1_chr11_+_129375841_129375863 | 0.72 | 4.2e-02 | Click! |
| HOXA3 | hg38_v1_chr7_-_27152561_27152598 | -0.30 | 4.8e-01 | Click! |
| DLX1 | hg38_v1_chr2_+_172084748_172084798 | 0.28 | 5.0e-01 | Click! |
Activity profile of DLX1_HOXA3_BARX2 motif
Sorted Z-values of DLX1_HOXA3_BARX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_41309434 | 1.10 |
ENST00000220772.8
|
SFRP1
|
secreted frizzled related protein 1 |
| chr1_+_160400543 | 0.90 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr1_+_27934980 | 0.86 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr3_-_151316795 | 0.75 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr10_-_104085847 | 0.69 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr3_-_191282383 | 0.68 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr18_+_36544544 | 0.67 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr1_-_242449478 | 0.66 |
ENST00000427495.5
|
PLD5
|
phospholipase D family member 5 |
| chr5_+_36608146 | 0.65 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr9_-_5304713 | 0.61 |
ENST00000381627.4
|
RLN2
|
relaxin 2 |
| chr3_-_112829367 | 0.60 |
ENST00000448932.4
ENST00000617549.3 |
CD200R1L
|
CD200 receptor 1 like |
| chr5_+_36606355 | 0.58 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr17_-_41118369 | 0.57 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr7_+_70596078 | 0.56 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr4_+_76435216 | 0.56 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr5_+_67004618 | 0.53 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_111999326 | 0.50 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr3_+_111999189 | 0.49 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr15_+_43693859 | 0.48 |
ENST00000413453.7
ENST00000415044.3 ENST00000626814.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr15_+_43594027 | 0.48 |
ENST00000453733.5
ENST00000441322.6 ENST00000627381.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr3_+_111998915 | 0.47 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr6_+_130018565 | 0.46 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr3_+_111998739 | 0.45 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr16_-_28623560 | 0.43 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr13_+_31739520 | 0.43 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr13_+_31739542 | 0.40 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr12_-_95116967 | 0.37 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr18_+_63587336 | 0.37 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13 |
| chr14_+_61187544 | 0.36 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr2_-_70553638 | 0.36 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr4_-_25863537 | 0.35 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr10_-_49762276 | 0.34 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr9_+_72577939 | 0.34 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr18_+_58221535 | 0.31 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr14_-_67412112 | 0.30 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr6_+_130421086 | 0.30 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A |
| chr17_-_7263959 | 0.30 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr9_-_5339874 | 0.29 |
ENST00000223862.2
|
RLN1
|
relaxin 1 |
| chr19_-_3557563 | 0.29 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr18_+_11689210 | 0.28 |
ENST00000334049.11
|
GNAL
|
G protein subunit alpha L |
| chr14_-_23183641 | 0.27 |
ENST00000469263.5
ENST00000525062.1 ENST00000316902.12 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 member 8 |
| chr1_+_196774813 | 0.27 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr2_-_101308681 | 0.27 |
ENST00000295317.4
|
RNF149
|
ring finger protein 149 |
| chr11_+_121576760 | 0.27 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr1_+_81306096 | 0.26 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr11_+_124241095 | 0.25 |
ENST00000641972.1
|
OR8G1
|
olfactory receptor family 8 subfamily G member 1 |
| chr13_+_36432487 | 0.25 |
ENST00000255465.7
ENST00000625767.1 |
CCNA1
|
cyclin A1 |
| chr8_-_85341705 | 0.25 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr10_-_49762335 | 0.25 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr13_-_85799400 | 0.24 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr6_+_125219804 | 0.24 |
ENST00000524679.1
|
TPD52L1
|
TPD52 like 1 |
| chr12_+_80716906 | 0.24 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr18_+_63587297 | 0.23 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr11_-_102780620 | 0.23 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr11_-_129024157 | 0.23 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr18_+_58341038 | 0.22 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr3_+_178419123 | 0.22 |
ENST00000614557.1
ENST00000455307.5 ENST00000436432.1 |
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript) long intergenic non-protein coding RNA 1014 |
| chr12_+_11649666 | 0.21 |
ENST00000396373.9
|
ETV6
|
ETS variant transcription factor 6 |
| chr3_-_112846856 | 0.21 |
ENST00000488794.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr11_-_5301946 | 0.21 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr4_-_73620391 | 0.20 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr17_-_40799939 | 0.20 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr3_-_161105399 | 0.20 |
ENST00000652593.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr1_+_76867469 | 0.20 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr4_+_87975829 | 0.20 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr12_-_21910853 | 0.20 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr6_+_121437378 | 0.19 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr9_+_12693327 | 0.19 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr4_+_87975667 | 0.19 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_207053229 | 0.19 |
ENST00000367080.8
ENST00000367079.3 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr2_+_227871618 | 0.19 |
ENST00000309931.3
ENST00000440997.1 |
DAW1
|
dynein assembly factor with WD repeats 1 |
| chr3_-_33645433 | 0.18 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr14_-_63728027 | 0.18 |
ENST00000247225.7
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr1_-_92486916 | 0.18 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr4_+_70334963 | 0.18 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr16_-_81220370 | 0.18 |
ENST00000337114.8
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chr3_-_161105224 | 0.18 |
ENST00000651254.1
ENST00000651178.1 ENST00000476999.6 ENST00000652596.1 ENST00000651305.1 ENST00000652111.1 ENST00000651292.1 ENST00000651282.1 ENST00000651380.1 ENST00000494173.7 ENST00000484127.5 ENST00000650733.1 ENST00000494818.6 ENST00000492353.5 ENST00000652143.1 ENST00000473142.5 ENST00000651147.1 ENST00000468268.5 ENST00000460353.2 ENST00000651953.1 ENST00000651972.1 ENST00000652730.1 ENST00000651460.1 ENST00000652059.1 ENST00000651509.1 ENST00000651801.1 ENST00000651686.1 ENST00000320474.10 ENST00000392781.7 ENST00000392779.6 ENST00000651791.1 ENST00000651117.1 ENST00000652032.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr1_-_197146688 | 0.18 |
ENST00000294732.11
|
ASPM
|
assembly factor for spindle microtubules |
| chr2_+_90038848 | 0.17 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr11_+_35180342 | 0.17 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr20_-_51802433 | 0.17 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr7_+_141776674 | 0.17 |
ENST00000247881.4
|
TAS2R4
|
taste 2 receptor member 4 |
| chr3_-_161105070 | 0.17 |
ENST00000651430.1
ENST00000650695.1 ENST00000651689.1 ENST00000651916.1 ENST00000488170.5 ENST00000652377.1 ENST00000652669.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr8_-_85341659 | 0.16 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr10_+_132397168 | 0.16 |
ENST00000631148.2
ENST00000305233.6 |
PWWP2B
|
PWWP domain containing 2B |
| chr12_-_10826358 | 0.16 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr7_-_111392915 | 0.16 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr21_+_42199686 | 0.16 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr13_-_75366973 | 0.16 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chr16_+_82056423 | 0.16 |
ENST00000568090.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr3_-_185821092 | 0.15 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr8_-_42377227 | 0.15 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr4_+_94974984 | 0.15 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr9_-_114348966 | 0.15 |
ENST00000374079.8
|
AKNA
|
AT-hook transcription factor |
| chr17_-_50707855 | 0.15 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr1_-_201171545 | 0.15 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr11_+_60280577 | 0.15 |
ENST00000679988.1
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr9_+_105700953 | 0.15 |
ENST00000374688.5
|
TMEM38B
|
transmembrane protein 38B |
| chr5_-_83673544 | 0.15 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_+_158968608 | 0.15 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr12_+_28257195 | 0.15 |
ENST00000381259.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr8_-_124565699 | 0.15 |
ENST00000519168.5
|
MTSS1
|
MTSS I-BAR domain containing 1 |
| chr3_-_98517096 | 0.15 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr1_+_154321107 | 0.15 |
ENST00000484864.1
|
AQP10
|
aquaporin 10 |
| chr11_-_107858777 | 0.15 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr14_+_56117702 | 0.14 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr20_-_51802509 | 0.14 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr21_-_30216047 | 0.14 |
ENST00000399899.2
|
CLDN8
|
claudin 8 |
| chr5_+_55853314 | 0.14 |
ENST00000354961.8
ENST00000297015.7 |
IL31RA
|
interleukin 31 receptor A |
| chr4_-_39977836 | 0.14 |
ENST00000303538.13
ENST00000503396.5 |
PDS5A
|
PDS5 cohesin associated factor A |
| chr6_+_26365159 | 0.14 |
ENST00000532865.5
ENST00000396934.7 ENST00000508906.6 ENST00000530653.5 ENST00000527417.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr11_-_117877463 | 0.14 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr9_+_128149447 | 0.14 |
ENST00000277480.7
ENST00000372998.1 |
LCN2
|
lipocalin 2 |
| chr20_+_33217325 | 0.14 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr10_-_14548646 | 0.14 |
ENST00000378470.5
|
FAM107B
|
family with sequence similarity 107 member B |
| chr8_-_42501224 | 0.14 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr2_-_133568393 | 0.14 |
ENST00000317721.10
ENST00000405974.7 ENST00000409261.6 ENST00000409213.5 |
NCKAP5
|
NCK associated protein 5 |
| chr6_+_26365176 | 0.14 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr2_+_230759918 | 0.14 |
ENST00000614925.1
|
CAB39
|
calcium binding protein 39 |
| chr11_-_117876719 | 0.14 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr10_-_133565542 | 0.14 |
ENST00000303903.10
ENST00000343131.7 |
SYCE1
|
synaptonemal complex central element protein 1 |
| chr6_+_26365215 | 0.13 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr13_-_110242694 | 0.13 |
ENST00000648989.1
ENST00000647797.1 ENST00000648966.1 ENST00000649484.1 ENST00000648695.1 ENST00000650115.1 ENST00000650566.1 |
COL4A1
|
collagen type IV alpha 1 chain |
| chr6_-_32941018 | 0.13 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_-_117876892 | 0.13 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr2_+_201182873 | 0.13 |
ENST00000360132.7
|
CASP10
|
caspase 10 |
| chr4_+_69931066 | 0.13 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr16_+_11965193 | 0.13 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr12_-_50222694 | 0.13 |
ENST00000552783.5
|
LIMA1
|
LIM domain and actin binding 1 |
| chr6_-_28443463 | 0.13 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr11_+_60280658 | 0.13 |
ENST00000337908.5
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr12_+_6904962 | 0.13 |
ENST00000415834.5
ENST00000436789.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr4_-_119322128 | 0.13 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr14_-_53956811 | 0.13 |
ENST00000559087.5
ENST00000245451.9 |
BMP4
|
bone morphogenetic protein 4 |
| chr12_+_20810698 | 0.13 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr11_-_57324907 | 0.13 |
ENST00000358252.8
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr17_-_41168219 | 0.13 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr17_-_66229380 | 0.13 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr7_+_143284930 | 0.13 |
ENST00000409244.5
ENST00000409541.5 ENST00000410004.1 ENST00000359333.8 |
TMEM139
|
transmembrane protein 139 |
| chr5_+_141177790 | 0.12 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr21_-_14210948 | 0.12 |
ENST00000681601.1
|
LIPI
|
lipase I |
| chr3_-_142029108 | 0.12 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr7_+_98106852 | 0.12 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr11_-_36598221 | 0.12 |
ENST00000311485.8
ENST00000527033.5 ENST00000532616.1 ENST00000618712.4 |
RAG2
|
recombination activating 2 |
| chr11_+_118527463 | 0.12 |
ENST00000302783.10
|
TTC36
|
tetratricopeptide repeat domain 36 |
| chr1_-_24415035 | 0.12 |
ENST00000374409.5
|
STPG1
|
sperm tail PG-rich repeat containing 1 |
| chr21_-_14210884 | 0.12 |
ENST00000679868.1
ENST00000400211.3 ENST00000680801.1 ENST00000536861.6 ENST00000614229.5 |
LIPI
|
lipase I |
| chr4_+_118888918 | 0.12 |
ENST00000434046.6
|
SYNPO2
|
synaptopodin 2 |
| chr8_+_104223320 | 0.12 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_-_53510445 | 0.12 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chr11_+_55827219 | 0.12 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor family 5 subfamily L member 2 |
| chr7_+_107583919 | 0.12 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr22_-_32464440 | 0.12 |
ENST00000397450.2
ENST00000397452.5 ENST00000300399.8 |
BPIFC
|
BPI fold containing family C |
| chrX_-_33211540 | 0.12 |
ENST00000357033.9
|
DMD
|
dystrophin |
| chr10_+_24208774 | 0.12 |
ENST00000376456.8
ENST00000458595.5 ENST00000376452.7 ENST00000430453.6 |
KIAA1217
|
KIAA1217 |
| chr16_-_28609992 | 0.12 |
ENST00000314752.11
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr2_+_90234809 | 0.12 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr11_-_129192291 | 0.12 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr14_+_69879408 | 0.12 |
ENST00000361956.8
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr6_+_29306626 | 0.12 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr11_-_129192198 | 0.12 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr11_+_65787056 | 0.11 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr4_-_73620629 | 0.11 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr5_+_90474848 | 0.11 |
ENST00000651687.1
|
POLR3G
|
RNA polymerase III subunit G |
| chr3_+_122055355 | 0.11 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr12_-_70637405 | 0.11 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr1_-_197146620 | 0.11 |
ENST00000367409.9
ENST00000680265.1 |
ASPM
|
assembly factor for spindle microtubules |
| chr5_+_90474879 | 0.11 |
ENST00000504930.5
ENST00000514483.5 |
POLR3G
|
RNA polymerase III subunit G |
| chr19_-_14979848 | 0.11 |
ENST00000594383.2
|
SLC1A6
|
solute carrier family 1 member 6 |
| chr3_-_194351290 | 0.11 |
ENST00000429275.1
ENST00000323830.4 |
CPN2
|
carboxypeptidase N subunit 2 |
| chr10_-_112183698 | 0.11 |
ENST00000369425.5
ENST00000348367.9 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr22_-_28711931 | 0.11 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr2_+_200889411 | 0.11 |
ENST00000409357.5
ENST00000409129.2 |
NIF3L1
|
NGG1 interacting factor 3 like 1 |
| chr11_+_112176364 | 0.11 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr17_-_75667165 | 0.11 |
ENST00000584999.1
ENST00000420326.6 ENST00000340830.9 |
RECQL5
|
RecQ like helicase 5 |
| chr8_+_97887903 | 0.11 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr15_+_58138368 | 0.11 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr21_-_30166782 | 0.11 |
ENST00000286808.5
|
CLDN17
|
claudin 17 |
| chr2_+_201183120 | 0.11 |
ENST00000272879.9
ENST00000286186.11 ENST00000374650.7 ENST00000346817.9 ENST00000313728.11 ENST00000448480.1 |
CASP10
|
caspase 10 |
| chr2_+_200889327 | 0.11 |
ENST00000426253.5
ENST00000454952.1 ENST00000651500.1 ENST00000409020.6 ENST00000359683.8 |
NIF3L1
|
NGG1 interacting factor 3 like 1 |
| chr3_+_4680617 | 0.11 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr19_+_9185594 | 0.11 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr6_+_116461364 | 0.10 |
ENST00000368606.7
ENST00000368605.3 |
CALHM6
|
calcium homeostasis modulator family member 6 |
| chrX_-_18672101 | 0.10 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr21_-_30881572 | 0.10 |
ENST00000332378.6
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr8_-_30812867 | 0.10 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr12_-_122500520 | 0.10 |
ENST00000540586.1
ENST00000543897.5 |
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr2_+_48314637 | 0.10 |
ENST00000413569.5
ENST00000340553.8 |
FOXN2
|
forkhead box N2 |
| chr8_+_104223344 | 0.10 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_+_130850585 | 0.10 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr14_+_22271921 | 0.10 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr13_-_35476682 | 0.10 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chr4_-_72569204 | 0.10 |
ENST00000286657.10
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif 3 |
| chr1_+_207325629 | 0.10 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr12_-_9869345 | 0.10 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr4_-_78939352 | 0.10 |
ENST00000512733.5
|
PAQR3
|
progestin and adipoQ receptor family member 3 |
| chr17_-_75667088 | 0.10 |
ENST00000578201.5
ENST00000423245.6 ENST00000317905.10 |
RECQL5
|
RecQ like helicase 5 |
| chr5_-_24644968 | 0.09 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr12_-_23584600 | 0.09 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr6_-_123636979 | 0.09 |
ENST00000662930.1
|
TRDN
|
triadin |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX1_HOXA3_BARX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.3 | 0.9 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 1.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.3 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.6 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.6 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.2 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.4 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:2000004 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0043017 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.0 | 0.2 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.8 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.2 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.0 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.1 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.0 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.3 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.0 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.0 | GO:0097183 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.5 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.0 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |