Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
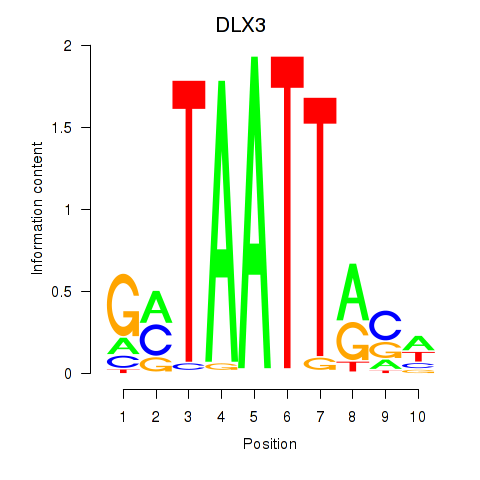
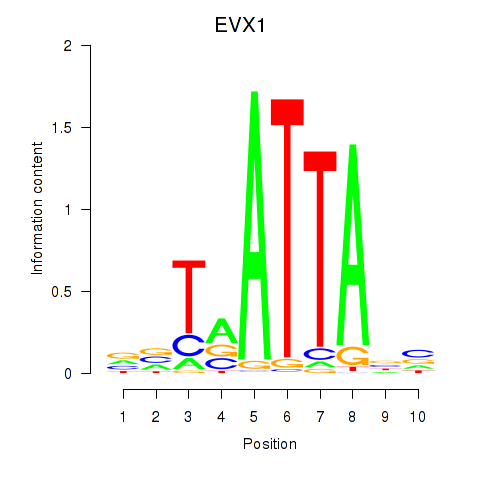
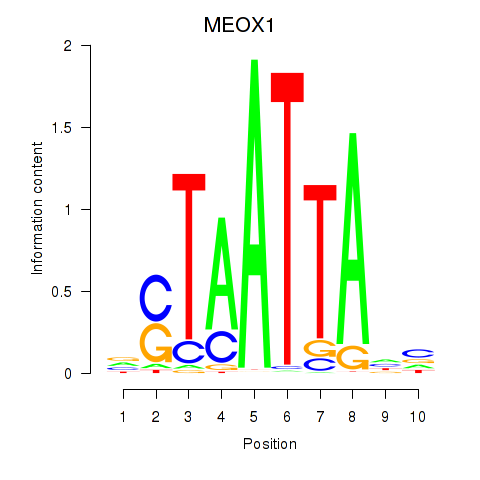
Results for DLX3_EVX1_MEOX1
Z-value: 0.17
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX3
|
ENSG00000064195.7 | distal-less homeobox 3 |
|
EVX1
|
ENSG00000106038.13 | even-skipped homeobox 1 |
|
MEOX1
|
ENSG00000005102.14 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EVX1 | hg38_v1_chr7_+_27242796_27242807 | -0.59 | 1.2e-01 | Click! |
| DLX3 | hg38_v1_chr17_-_49995210_49995230 | 0.38 | 3.6e-01 | Click! |
| MEOX1 | hg38_v1_chr17_-_43661915_43661929 | -0.28 | 5.0e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_70596078 | 0.39 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr1_+_160190567 | 0.38 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr5_+_67004618 | 0.36 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_116427839 | 0.33 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr12_+_107318395 | 0.31 |
ENST00000420571.6
ENST00000280758.10 |
BTBD11
|
BTB domain containing 11 |
| chr6_+_29306626 | 0.28 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr8_-_42377227 | 0.28 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr18_-_36129305 | 0.26 |
ENST00000269187.10
ENST00000590986.5 ENST00000440549.6 |
SLC39A6
|
solute carrier family 39 member 6 |
| chr4_+_76435216 | 0.26 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr6_+_130018565 | 0.25 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr3_+_111999326 | 0.22 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr3_+_111998915 | 0.21 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr20_+_59835853 | 0.21 |
ENST00000492611.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr3_+_111999189 | 0.20 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr1_-_93681829 | 0.20 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr2_+_168901290 | 0.18 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr3_+_111998739 | 0.18 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr14_+_22271921 | 0.17 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr10_-_104085847 | 0.16 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr4_+_40196907 | 0.15 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr4_-_65670478 | 0.15 |
ENST00000613740.5
ENST00000622150.4 ENST00000511294.1 |
EPHA5
|
EPH receptor A5 |
| chr13_-_46142834 | 0.15 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr7_+_117020191 | 0.15 |
ENST00000434836.5
ENST00000393443.5 ENST00000465133.5 ENST00000477742.5 ENST00000393444.7 ENST00000393447.8 |
ST7
|
suppression of tumorigenicity 7 |
| chr7_-_101217569 | 0.14 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr22_-_32464440 | 0.14 |
ENST00000397450.2
ENST00000397452.5 ENST00000300399.8 |
BPIFC
|
BPI fold containing family C |
| chr16_+_24729692 | 0.14 |
ENST00000315183.11
|
TNRC6A
|
trinucleotide repeat containing adaptor 6A |
| chr1_+_13585453 | 0.13 |
ENST00000487038.5
ENST00000475043.5 |
PDPN
|
podoplanin |
| chr4_-_65670339 | 0.13 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5 |
| chr17_-_7263959 | 0.12 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr6_+_106360668 | 0.12 |
ENST00000633556.3
|
CRYBG1
|
crystallin beta-gamma domain containing 1 |
| chr1_+_160400543 | 0.12 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr15_+_89088417 | 0.12 |
ENST00000569550.5
ENST00000565066.5 ENST00000565973.5 ENST00000352732.10 |
ABHD2
|
abhydrolase domain containing 2, acylglycerol lipase |
| chr6_+_47698574 | 0.12 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr6_+_47698538 | 0.12 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr15_+_74788542 | 0.12 |
ENST00000567571.5
|
CSK
|
C-terminal Src kinase |
| chr20_+_61599755 | 0.12 |
ENST00000543233.2
|
CDH4
|
cadherin 4 |
| chr4_-_115113822 | 0.12 |
ENST00000613194.4
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr4_+_40197023 | 0.11 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr16_+_69105636 | 0.11 |
ENST00000569188.6
|
HAS3
|
hyaluronan synthase 3 |
| chr18_+_34593392 | 0.11 |
ENST00000684377.1
|
DTNA
|
dystrobrevin alpha |
| chr11_+_65787056 | 0.11 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr19_-_3557563 | 0.11 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr5_-_16508858 | 0.11 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1 |
| chrX_+_30235894 | 0.11 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3 |
| chr5_-_16508951 | 0.10 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr1_+_158461574 | 0.10 |
ENST00000641432.1
ENST00000641460.1 ENST00000641535.1 ENST00000641971.1 |
OR10K1
|
olfactory receptor family 10 subfamily K member 1 |
| chr5_-_16508812 | 0.10 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr7_-_33062750 | 0.10 |
ENST00000610140.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr2_-_181680490 | 0.10 |
ENST00000684145.1
ENST00000295108.4 ENST00000684079.1 ENST00000683430.1 |
CERKL
NEUROD1
|
ceramide kinase like neuronal differentiation 1 |
| chr12_-_89352487 | 0.10 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr6_+_151325665 | 0.10 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr10_+_116324440 | 0.09 |
ENST00000333254.4
|
CCDC172
|
coiled-coil domain containing 172 |
| chr20_-_51802433 | 0.09 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr1_+_236795254 | 0.09 |
ENST00000366577.10
ENST00000674797.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr1_-_68232514 | 0.09 |
ENST00000262348.9
ENST00000370973.2 ENST00000370971.1 |
WLS
|
Wnt ligand secretion mediator |
| chr12_-_89352395 | 0.09 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr21_-_30881572 | 0.09 |
ENST00000332378.6
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr11_-_124320197 | 0.08 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chrX_-_18672101 | 0.08 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr8_+_32721823 | 0.08 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr5_+_36606355 | 0.08 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr19_+_12938598 | 0.08 |
ENST00000586760.2
ENST00000316448.10 ENST00000588454.6 |
CALR
|
calreticulin |
| chrM_+_12329 | 0.08 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr3_-_151316795 | 0.08 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_-_24644968 | 0.08 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr5_-_16508788 | 0.08 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr3_-_123620496 | 0.08 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr17_-_41149823 | 0.08 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr5_-_16508990 | 0.08 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1 |
| chr18_+_58221535 | 0.08 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr13_-_35476682 | 0.08 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chr1_-_68232539 | 0.08 |
ENST00000370976.7
ENST00000354777.6 |
WLS
|
Wnt ligand secretion mediator |
| chr5_+_127649018 | 0.07 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr1_+_99646025 | 0.07 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr4_-_138242325 | 0.07 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr14_+_64704380 | 0.07 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr1_-_152414256 | 0.07 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr11_-_129192198 | 0.07 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr8_-_42501224 | 0.07 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr14_+_61187544 | 0.07 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr18_+_58341038 | 0.07 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr12_+_41437680 | 0.07 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr4_+_173168800 | 0.07 |
ENST00000512285.5
ENST00000265000.9 |
GALNT7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr9_+_90827452 | 0.07 |
ENST00000375746.1
|
SYK
|
spleen associated tyrosine kinase |
| chr22_-_33572227 | 0.06 |
ENST00000674780.1
|
LARGE1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr10_+_18400562 | 0.06 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr1_-_197146620 | 0.06 |
ENST00000367409.9
ENST00000680265.1 |
ASPM
|
assembly factor for spindle microtubules |
| chr18_+_34593312 | 0.06 |
ENST00000591816.6
ENST00000588125.5 ENST00000684610.1 ENST00000683705.1 ENST00000598334.5 ENST00000588684.5 ENST00000554864.7 ENST00000399121.9 ENST00000595022.5 |
DTNA
|
dystrobrevin alpha |
| chr18_+_78979811 | 0.06 |
ENST00000537592.7
|
SALL3
|
spalt like transcription factor 3 |
| chrX_-_100874332 | 0.06 |
ENST00000372960.8
|
NOX1
|
NADPH oxidase 1 |
| chr7_+_130266847 | 0.06 |
ENST00000222481.9
|
CPA2
|
carboxypeptidase A2 |
| chr7_-_93226449 | 0.06 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr3_-_74521140 | 0.06 |
ENST00000263665.6
|
CNTN3
|
contactin 3 |
| chr1_-_183653307 | 0.06 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr15_+_41621492 | 0.06 |
ENST00000570161.6
|
MGA
|
MAX dimerization protein MGA |
| chr1_+_202348727 | 0.06 |
ENST00000356764.6
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chrM_+_8489 | 0.06 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase membrane subunit 6 |
| chr20_-_51802509 | 0.06 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr3_+_130850585 | 0.06 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr2_+_112542413 | 0.06 |
ENST00000417433.6
ENST00000263331.10 |
POLR1B
|
RNA polymerase I subunit B |
| chrX_-_19670983 | 0.06 |
ENST00000379716.5
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr17_-_40755328 | 0.06 |
ENST00000312150.5
|
KRT25
|
keratin 25 |
| chr11_-_107858777 | 0.06 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr11_-_129192291 | 0.06 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr4_-_39977836 | 0.06 |
ENST00000303538.13
ENST00000503396.5 |
PDS5A
|
PDS5 cohesin associated factor A |
| chr17_-_66229380 | 0.06 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chrX_-_100874351 | 0.06 |
ENST00000372966.8
|
NOX1
|
NADPH oxidase 1 |
| chr6_+_156776020 | 0.06 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B |
| chr5_-_169980474 | 0.06 |
ENST00000377365.4
|
INSYN2B
|
inhibitory synaptic factor family member 2B |
| chr14_-_81533800 | 0.06 |
ENST00000555824.5
ENST00000557372.1 ENST00000336735.9 |
SEL1L
|
SEL1L adaptor subunit of ERAD E3 ubiquitin ligase |
| chr15_+_41621134 | 0.05 |
ENST00000566718.6
|
MGA
|
MAX dimerization protein MGA |
| chr11_+_20599602 | 0.05 |
ENST00000525748.6
|
SLC6A5
|
solute carrier family 6 member 5 |
| chr17_-_40799939 | 0.05 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr21_-_41926680 | 0.05 |
ENST00000329623.11
|
C2CD2
|
C2 calcium dependent domain containing 2 |
| chr7_-_32299287 | 0.05 |
ENST00000396193.5
|
PDE1C
|
phosphodiesterase 1C |
| chr14_+_64214136 | 0.05 |
ENST00000557084.1
ENST00000458046.6 |
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr3_+_44799187 | 0.05 |
ENST00000425755.5
|
KIF15
|
kinesin family member 15 |
| chr13_+_73058993 | 0.05 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5 |
| chr17_+_44187190 | 0.05 |
ENST00000319511.6
|
TMUB2
|
transmembrane and ubiquitin like domain containing 2 |
| chr17_+_44187210 | 0.05 |
ENST00000589785.1
ENST00000592825.1 ENST00000589184.5 |
TMUB2
|
transmembrane and ubiquitin like domain containing 2 |
| chr10_+_24449426 | 0.05 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217 |
| chr3_-_123961399 | 0.05 |
ENST00000488653.6
|
CCDC14
|
coiled-coil domain containing 14 |
| chr12_-_10826358 | 0.05 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chrX_+_136648138 | 0.05 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chrX_+_106611930 | 0.05 |
ENST00000372544.6
ENST00000372548.9 |
RADX
|
RPA1 related single stranded DNA binding protein, X-linked |
| chrX_-_21658324 | 0.05 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr7_+_99828010 | 0.05 |
ENST00000631161.2
ENST00000354829.7 ENST00000342499.8 ENST00000417625.5 ENST00000415413.5 ENST00000444905.5 ENST00000222382.5 ENST00000312017.9 |
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43 |
| chr16_+_76277568 | 0.05 |
ENST00000622250.4
|
CNTNAP4
|
contactin associated protein family member 4 |
| chr14_+_32329256 | 0.05 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr15_-_43106022 | 0.05 |
ENST00000627960.1
ENST00000290650.9 |
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr14_+_69879408 | 0.05 |
ENST00000361956.8
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr11_-_120138104 | 0.05 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr3_-_108529322 | 0.05 |
ENST00000273353.4
|
MYH15
|
myosin heavy chain 15 |
| chr12_-_119803383 | 0.05 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr20_+_33079640 | 0.04 |
ENST00000375483.4
|
BPIFB4
|
BPI fold containing family B member 4 |
| chr7_-_81770122 | 0.04 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr2_-_207167220 | 0.04 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr12_-_6124662 | 0.04 |
ENST00000261405.10
|
VWF
|
von Willebrand factor |
| chr9_-_123184233 | 0.04 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr8_-_102412686 | 0.04 |
ENST00000220959.8
ENST00000520539.6 |
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr3_-_33645433 | 0.04 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr7_-_73624492 | 0.04 |
ENST00000414749.6
ENST00000429400.6 ENST00000434326.5 ENST00000313375.8 ENST00000354613.5 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein like |
| chr15_-_31101707 | 0.04 |
ENST00000397795.6
ENST00000256552.11 ENST00000559179.2 |
TRPM1
|
transient receptor potential cation channel subfamily M member 1 |
| chr4_+_69931066 | 0.04 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr15_-_68205319 | 0.04 |
ENST00000467889.3
ENST00000448060.7 |
CALML4
|
calmodulin like 4 |
| chr7_+_154305256 | 0.04 |
ENST00000619756.4
|
DPP6
|
dipeptidyl peptidase like 6 |
| chr10_+_84194527 | 0.04 |
ENST00000623527.4
|
CDHR1
|
cadherin related family member 1 |
| chr5_+_98773651 | 0.04 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr4_+_87832917 | 0.04 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_-_25863537 | 0.04 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr12_-_56300299 | 0.04 |
ENST00000552688.5
ENST00000548041.5 ENST00000551137.5 ENST00000551968.5 ENST00000351328.8 ENST00000542324.6 ENST00000546930.5 ENST00000549221.5 ENST00000550159.5 ENST00000550734.5 |
CS
|
citrate synthase |
| chr12_-_21775581 | 0.04 |
ENST00000537950.1
ENST00000665145.1 |
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr4_-_173530219 | 0.04 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr1_-_201171545 | 0.04 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr18_+_63542365 | 0.04 |
ENST00000269491.6
ENST00000382768.2 |
SERPINB12
|
serpin family B member 12 |
| chr3_-_69080350 | 0.04 |
ENST00000630585.1
ENST00000361055.9 ENST00000415609.6 ENST00000349511.8 |
UBA3
|
ubiquitin like modifier activating enzyme 3 |
| chr4_-_115113614 | 0.04 |
ENST00000264363.7
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr12_-_56300358 | 0.04 |
ENST00000550655.5
ENST00000548567.5 ENST00000551430.6 |
CS
|
citrate synthase |
| chrX_+_134796758 | 0.04 |
ENST00000414371.6
|
PABIR3
|
PABIR family member 3 |
| chrX_-_108736556 | 0.04 |
ENST00000372129.4
|
IRS4
|
insulin receptor substrate 4 |
| chr11_-_122116215 | 0.04 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr15_-_68205274 | 0.04 |
ENST00000540479.6
|
CALML4
|
calmodulin like 4 |
| chr15_+_88621290 | 0.04 |
ENST00000332810.4
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr3_-_114179052 | 0.03 |
ENST00000383673.5
ENST00000295881.9 |
DRD3
|
dopamine receptor D3 |
| chr16_+_24729641 | 0.03 |
ENST00000395799.8
|
TNRC6A
|
trinucleotide repeat containing adaptor 6A |
| chr8_-_109334074 | 0.03 |
ENST00000239690.9
|
NUDCD1
|
NudC domain containing 1 |
| chr22_-_41286168 | 0.03 |
ENST00000356244.8
|
RANGAP1
|
Ran GTPase activating protein 1 |
| chr12_-_14696571 | 0.03 |
ENST00000261170.5
|
GUCY2C
|
guanylate cyclase 2C |
| chr8_-_86743626 | 0.03 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3 |
| chr6_+_31927486 | 0.03 |
ENST00000442278.6
|
C2
|
complement C2 |
| chr14_+_56117702 | 0.03 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr18_-_55587335 | 0.03 |
ENST00000638154.3
|
TCF4
|
transcription factor 4 |
| chr3_+_124384513 | 0.03 |
ENST00000682540.1
ENST00000522553.6 ENST00000682695.1 ENST00000682674.1 ENST00000684382.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr8_-_109334112 | 0.03 |
ENST00000678094.1
|
NUDCD1
|
NudC domain containing 1 |
| chr2_+_27275429 | 0.03 |
ENST00000420191.5
ENST00000296097.8 |
DNAJC5G
|
DnaJ heat shock protein family (Hsp40) member C5 gamma |
| chrX_-_107118783 | 0.03 |
ENST00000372487.5
ENST00000372479.7 |
RBM41
|
RNA binding motif protein 41 |
| chr2_-_17800195 | 0.03 |
ENST00000402989.5
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr14_-_21269451 | 0.03 |
ENST00000336053.10
|
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C |
| chr7_-_122304738 | 0.03 |
ENST00000442488.7
|
FEZF1
|
FEZ family zinc finger 1 |
| chr19_-_14979848 | 0.03 |
ENST00000594383.2
|
SLC1A6
|
solute carrier family 1 member 6 |
| chr16_+_7303245 | 0.03 |
ENST00000674626.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr15_-_34583592 | 0.03 |
ENST00000683415.1
|
GOLGA8B
|
golgin A8 family member B |
| chr19_+_4007714 | 0.03 |
ENST00000262971.3
|
PIAS4
|
protein inhibitor of activated STAT 4 |
| chrM_+_7586 | 0.03 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr3_+_4680617 | 0.03 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr8_-_102412740 | 0.03 |
ENST00000521922.5
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr20_+_18507520 | 0.03 |
ENST00000336714.8
ENST00000646240.1 ENST00000450074.6 ENST00000262544.6 |
SEC23B
|
SEC23 homolog B, COPII coat complex component |
| chr6_-_75493629 | 0.03 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1 |
| chr9_+_121567057 | 0.03 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr12_-_55296569 | 0.03 |
ENST00000358433.3
|
OR6C6
|
olfactory receptor family 6 subfamily C member 6 |
| chr16_-_67483541 | 0.03 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr2_+_48314637 | 0.03 |
ENST00000413569.5
ENST00000340553.8 |
FOXN2
|
forkhead box N2 |
| chr7_-_25228485 | 0.03 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr6_+_157036315 | 0.03 |
ENST00000637904.1
|
ARID1B
|
AT-rich interaction domain 1B |
| chr5_-_88883420 | 0.03 |
ENST00000437473.6
|
MEF2C
|
myocyte enhancer factor 2C |
| chr9_+_127264740 | 0.03 |
ENST00000373387.9
|
GARNL3
|
GTPase activating Rap/RanGAP domain like 3 |
| chr8_+_7926337 | 0.03 |
ENST00000400120.3
|
ZNF705B
|
zinc finger protein 705B |
| chr12_-_42484298 | 0.02 |
ENST00000640055.1
ENST00000639566.1 ENST00000455697.6 ENST00000639589.1 |
PRICKLE1
|
prickle planar cell polarity protein 1 |
| chr7_-_122304499 | 0.02 |
ENST00000427185.2
|
FEZF1
|
FEZ family zinc finger 1 |
| chr6_-_109381739 | 0.02 |
ENST00000504373.2
|
CD164
|
CD164 molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX3_EVX1_MEOX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.4 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.4 | GO:0014894 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |