Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
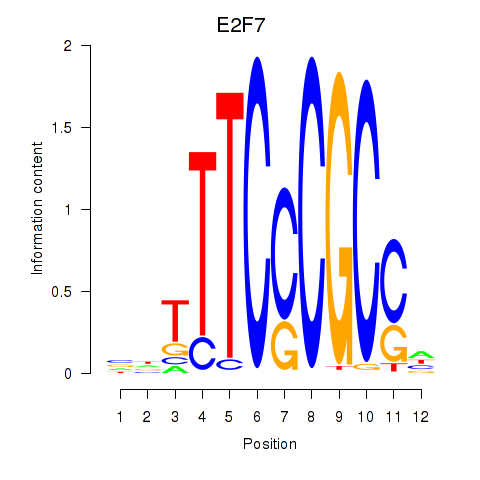
Results for E2F7_E2F1
Z-value: 0.17
Transcription factors associated with E2F7_E2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F7
|
ENSG00000165891.16 | E2F transcription factor 7 |
|
E2F1
|
ENSG00000101412.13 | E2F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F7 | hg38_v1_chr12_-_77065526_77065586 | 0.93 | 7.7e-04 | Click! |
| E2F1 | hg38_v1_chr20_-_33686371_33686407 | 0.52 | 1.9e-01 | Click! |
Activity profile of E2F7_E2F1 motif
Sorted Z-values of E2F7_E2F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F7_E2F1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.7 | 2.1 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.4 | 2.2 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.3 | 1.0 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.3 | 3.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 1.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 0.8 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 2.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.0 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 0.7 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.2 | 0.7 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.2 | 0.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.2 | 0.8 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.2 | 0.6 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.6 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.2 | 0.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 1.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 1.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.2 | 0.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 1.0 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 0.8 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 | 0.6 | GO:2000685 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) oncogene-induced cell senescence(GO:0090402) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.2 | 0.8 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.2 | 0.5 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.7 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.6 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.4 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.4 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.6 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.3 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 0.1 | 0.3 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.4 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.5 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.1 | 0.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.6 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 0.4 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.5 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.5 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.3 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 1.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.4 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.5 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.3 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.3 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.4 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 0.1 | GO:0035739 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.1 | 1.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.9 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.5 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.7 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 0.8 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.6 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 1.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 2.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.3 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.2 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.6 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.8 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.4 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 0.2 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.2 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) telomerase RNA stabilization(GO:0090669) |
| 0.1 | 2.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.1 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.2 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.2 | GO:1990834 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.1 | 0.2 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.4 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.2 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.2 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 2.2 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.2 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.1 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.2 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.8 | GO:0045005 | DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.1 | 0.2 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.7 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.8 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.2 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.1 | 0.3 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 1.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 0.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.5 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0051821 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) development involved in symbiotic interaction(GO:0044111) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.0 | 0.6 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.4 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.1 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 0.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.5 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.2 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.2 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.1 | GO:0031445 | regulation of heterochromatin assembly(GO:0031445) positive regulation of heterochromatin assembly(GO:0031453) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.6 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0003285 | septum secundum development(GO:0003285) atrial septum secundum morphogenesis(GO:0003290) |
| 0.0 | 0.6 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.6 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.2 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 0.0 | 0.3 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.8 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.0 | 0.5 | GO:0090220 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.1 | GO:0072209 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.2 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.0 | 0.2 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.2 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:1905216 | positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1902908 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 1.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0042853 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 1.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.3 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 0.1 | GO:0007314 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.0 | 0.3 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0034414 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0061573 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.6 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:1904798 | mesodermal cell fate determination(GO:0007500) negative regulation of muscle hyperplasia(GO:0014740) positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.0 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.3 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.2 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0003264 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0021722 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:1903891 | regulation of ATF6-mediated unfolded protein response(GO:1903891) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.8 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.6 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 1.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) negative regulation of mesodermal cell fate specification(GO:0042662) |
| 0.0 | 0.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.3 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.3 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.0 | GO:0070256 | negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0021502 | neural fold elevation formation(GO:0021502) nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.0 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.3 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.1 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.6 | 2.8 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.4 | 1.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 1.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 0.8 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.3 | 2.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 1.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 2.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.7 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.2 | 1.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.3 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 0.4 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 0.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.6 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 1.0 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.1 | 0.4 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 1.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.5 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 2.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.4 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.9 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 1.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.1 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.1 | GO:0090576 | transcription factor TFIIIC complex(GO:0000127) RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 2.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 2.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 0.8 | GO:0016420 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.2 | 0.7 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 2.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.6 | GO:0004638 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 0.8 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 0.6 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.6 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.2 | 0.6 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.7 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.7 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.6 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.5 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 4.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 1.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.3 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.4 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.3 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 1.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.2 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.6 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.3 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.3 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.1 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.2 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.1 | 0.2 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 1.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.2 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 0.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.2 | GO:0032143 | single guanine insertion binding(GO:0032142) single thymine insertion binding(GO:0032143) |
| 0.1 | 0.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 1.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004487 | formate-tetrahydrofolate ligase activity(GO:0004329) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.4 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 2.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 1.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.6 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 1.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 2.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0033749 | histone demethylase activity (H3-R2 specific)(GO:0033746) histone demethylase activity (H4-R3 specific)(GO:0033749) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 6.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 5.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 3.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 3.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.1 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.4 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 1.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 2.0 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |