Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
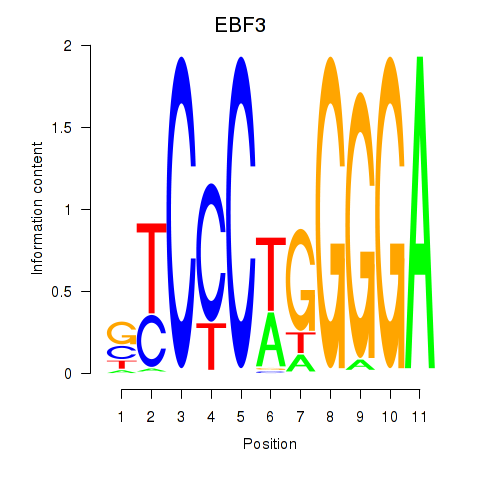
Results for EBF3
Z-value: 0.29
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.16 | EBF transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EBF3 | hg38_v1_chr10_-_129964240_129964291 | 0.39 | 3.4e-01 | Click! |
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_21536884 | 0.33 |
ENST00000546363.5
|
SALL2
|
spalt like transcription factor 2 |
| chr16_-_68236069 | 0.29 |
ENST00000473183.7
ENST00000565858.5 |
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr20_+_59835853 | 0.26 |
ENST00000492611.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr16_-_85750951 | 0.23 |
ENST00000602675.5
|
C16orf74
|
chromosome 16 open reading frame 74 |
| chr16_-_85751112 | 0.21 |
ENST00000602766.1
|
C16orf74
|
chromosome 16 open reading frame 74 |
| chr16_-_85751028 | 0.20 |
ENST00000284245.9
ENST00000602914.1 |
C16orf74
|
chromosome 16 open reading frame 74 |
| chr19_-_35513641 | 0.20 |
ENST00000339686.8
ENST00000447113.6 |
DMKN
|
dermokine |
| chr19_-_38253238 | 0.19 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr16_+_67199104 | 0.19 |
ENST00000360833.6
ENST00000652269.1 ENST00000393997.8 |
ELMO3
|
engulfment and cell motility 3 |
| chr3_-_190122317 | 0.18 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr5_-_176537361 | 0.17 |
ENST00000274811.9
|
RNF44
|
ring finger protein 44 |
| chr1_+_34755039 | 0.17 |
ENST00000338513.1
|
GJB5
|
gap junction protein beta 5 |
| chr4_+_1793285 | 0.15 |
ENST00000440486.8
ENST00000412135.7 ENST00000481110.7 ENST00000340107.8 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr1_-_153544997 | 0.15 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chrX_+_136205982 | 0.15 |
ENST00000628568.1
|
FHL1
|
four and a half LIM domains 1 |
| chr1_+_153357846 | 0.15 |
ENST00000368738.4
|
S100A9
|
S100 calcium binding protein A9 |
| chr8_+_22579139 | 0.14 |
ENST00000397761.6
|
PDLIM2
|
PDZ and LIM domain 2 |
| chr8_+_22578735 | 0.14 |
ENST00000339162.11
ENST00000308354.11 |
PDLIM2
|
PDZ and LIM domain 2 |
| chr1_+_26543106 | 0.14 |
ENST00000530003.5
|
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr14_+_24398986 | 0.14 |
ENST00000382554.4
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr2_-_237590694 | 0.14 |
ENST00000264601.8
ENST00000411462.5 ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr15_+_73926443 | 0.13 |
ENST00000261921.8
|
LOXL1
|
lysyl oxidase like 1 |
| chr17_-_58417521 | 0.13 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr17_-_58417547 | 0.13 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43 |
| chr2_-_237590660 | 0.13 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr8_+_22579100 | 0.13 |
ENST00000452226.5
ENST00000397760.8 |
PDLIM2
|
PDZ and LIM domain 2 |
| chr2_-_46542555 | 0.13 |
ENST00000522587.6
|
ATP6V1E2
|
ATPase H+ transporting V1 subunit E2 |
| chrX_-_48971829 | 0.12 |
ENST00000218176.4
|
KCND1
|
potassium voltage-gated channel subfamily D member 1 |
| chr17_-_41521719 | 0.12 |
ENST00000393976.6
|
KRT15
|
keratin 15 |
| chr1_-_159945596 | 0.12 |
ENST00000361509.7
ENST00000611023.1 ENST00000368094.6 |
IGSF9
|
immunoglobulin superfamily member 9 |
| chr4_-_89837076 | 0.12 |
ENST00000506691.1
|
SNCA
|
synuclein alpha |
| chr11_+_1839452 | 0.12 |
ENST00000381906.5
|
TNNI2
|
troponin I2, fast skeletal type |
| chr18_+_11981488 | 0.12 |
ENST00000269159.8
|
IMPA2
|
inositol monophosphatase 2 |
| chr1_+_2073462 | 0.12 |
ENST00000400921.6
|
PRKCZ
|
protein kinase C zeta |
| chr17_-_7590072 | 0.11 |
ENST00000538513.6
ENST00000570788.1 ENST00000250055.3 |
SOX15
|
SRY-box transcription factor 15 |
| chrX_+_136147465 | 0.11 |
ENST00000651929.2
|
FHL1
|
four and a half LIM domains 1 |
| chr11_+_1839602 | 0.11 |
ENST00000617947.4
ENST00000252898.11 |
TNNI2
|
troponin I2, fast skeletal type |
| chr1_+_2073986 | 0.11 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr19_+_46602050 | 0.11 |
ENST00000599839.5
ENST00000596362.1 |
CALM3
|
calmodulin 3 |
| chr4_-_89837106 | 0.11 |
ENST00000394986.5
ENST00000394991.8 ENST00000506244.5 ENST00000394989.6 ENST00000673902.1 ENST00000674129.1 |
SNCA
|
synuclein alpha |
| chr2_+_27078598 | 0.11 |
ENST00000380320.9
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr9_-_114348966 | 0.11 |
ENST00000374079.8
|
AKNA
|
AT-hook transcription factor |
| chr7_-_151814668 | 0.11 |
ENST00000651764.1
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr16_+_67199509 | 0.11 |
ENST00000477898.5
|
ELMO3
|
engulfment and cell motility 3 |
| chrX_+_136147525 | 0.11 |
ENST00000652745.1
ENST00000627578.2 ENST00000652457.1 ENST00000394155.8 ENST00000618438.4 |
FHL1
|
four and a half LIM domains 1 |
| chr12_-_86838867 | 0.11 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C |
| chrX_+_136147556 | 0.11 |
ENST00000651089.1
ENST00000420362.5 |
FHL1
|
four and a half LIM domains 1 |
| chr1_-_116667668 | 0.11 |
ENST00000369486.8
ENST00000369483.5 |
IGSF3
|
immunoglobulin superfamily member 3 |
| chr5_+_10564064 | 0.11 |
ENST00000296657.7
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr1_+_32765667 | 0.10 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr1_+_1033987 | 0.10 |
ENST00000651234.1
ENST00000652369.1 |
AGRN
|
agrin |
| chr11_+_1838970 | 0.10 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type |
| chr19_+_33373694 | 0.10 |
ENST00000284000.9
|
CEBPG
|
CCAAT enhancer binding protein gamma |
| chr11_+_64234569 | 0.10 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr2_-_64144411 | 0.10 |
ENST00000358912.5
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chr10_+_97572771 | 0.10 |
ENST00000370655.6
ENST00000455090.1 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr10_+_97572493 | 0.10 |
ENST00000307518.9
ENST00000298808.9 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr19_+_33374312 | 0.10 |
ENST00000585933.2
|
CEBPG
|
CCAAT enhancer binding protein gamma |
| chr4_-_86594037 | 0.10 |
ENST00000641050.1
ENST00000641831.1 ENST00000515400.3 ENST00000641391.1 ENST00000641157.1 ENST00000641737.1 ENST00000502302.6 ENST00000640527.1 ENST00000512046.2 ENST00000513186.7 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_+_22054817 | 0.10 |
ENST00000432128.5
ENST00000443491.6 ENST00000517600.5 ENST00000523782.6 |
DMTN
|
dematin actin binding protein |
| chr17_-_7393404 | 0.10 |
ENST00000575434.4
|
PLSCR3
|
phospholipid scramblase 3 |
| chr19_+_15049469 | 0.10 |
ENST00000427043.4
|
CASP14
|
caspase 14 |
| chr18_+_36544544 | 0.10 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr8_-_109608055 | 0.09 |
ENST00000529690.5
|
SYBU
|
syntabulin |
| chr19_-_4338786 | 0.09 |
ENST00000601482.1
ENST00000600324.5 ENST00000594605.6 |
STAP2
|
signal transducing adaptor family member 2 |
| chr16_+_23557714 | 0.09 |
ENST00000567212.5
ENST00000567264.1 ENST00000395878.8 |
UBFD1
|
ubiquitin family domain containing 1 |
| chr17_-_75878542 | 0.09 |
ENST00000254816.6
|
TRIM47
|
tripartite motif containing 47 |
| chr2_+_42568510 | 0.09 |
ENST00000407270.7
|
MTA3
|
metastasis associated 1 family member 3 |
| chr17_+_7404851 | 0.09 |
ENST00000575301.5
|
NLGN2
|
neuroligin 2 |
| chr7_+_107470050 | 0.09 |
ENST00000304402.6
|
GPR22
|
G protein-coupled receptor 22 |
| chr7_-_151814636 | 0.09 |
ENST00000652047.1
ENST00000650858.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chrX_+_48786578 | 0.09 |
ENST00000376670.9
|
GATA1
|
GATA binding protein 1 |
| chr1_-_153550083 | 0.09 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr7_-_151814819 | 0.08 |
ENST00000392801.6
ENST00000652707.1 ENST00000651378.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chrX_-_33211540 | 0.08 |
ENST00000357033.9
|
DMD
|
dystrophin |
| chr6_-_106974721 | 0.08 |
ENST00000606017.2
ENST00000620405.1 |
CD24
|
CD24 molecule |
| chr7_-_151877105 | 0.08 |
ENST00000287878.9
ENST00000652321.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr2_+_219461255 | 0.08 |
ENST00000396686.5
ENST00000396689.2 |
SPEG
|
striated muscle enriched protein kinase |
| chr1_-_42817357 | 0.08 |
ENST00000372521.9
|
SVBP
|
small vasohibin binding protein |
| chr2_+_219460719 | 0.08 |
ENST00000396688.5
|
SPEG
|
striated muscle enriched protein kinase |
| chr14_+_22495890 | 0.08 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr11_-_72721908 | 0.08 |
ENST00000426523.5
ENST00000429686.5 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_-_74507664 | 0.08 |
ENST00000233630.11
|
PCGF1
|
polycomb group ring finger 1 |
| chr9_+_130200375 | 0.08 |
ENST00000630865.1
|
NCS1
|
neuronal calcium sensor 1 |
| chr1_-_6419903 | 0.07 |
ENST00000377836.8
ENST00000487437.5 ENST00000489730.1 ENST00000377834.8 |
HES2
|
hes family bHLH transcription factor 2 |
| chr12_+_50085194 | 0.07 |
ENST00000381513.8
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr17_+_42289213 | 0.07 |
ENST00000677301.1
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr12_+_50085325 | 0.07 |
ENST00000551966.5
ENST00000550477.5 ENST00000394963.9 |
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr18_-_51197671 | 0.07 |
ENST00000406189.4
|
MEX3C
|
mex-3 RNA binding family member C |
| chr16_+_81444799 | 0.07 |
ENST00000537098.8
|
CMIP
|
c-Maf inducing protein |
| chr11_+_20363685 | 0.07 |
ENST00000530266.5
ENST00000451739.7 ENST00000421577.6 ENST00000443524.6 ENST00000419348.6 |
HTATIP2
|
HIV-1 Tat interactive protein 2 |
| chr22_+_37805218 | 0.07 |
ENST00000340857.4
|
H1-0
|
H1.0 linker histone |
| chrX_+_48786562 | 0.07 |
ENST00000651144.1
ENST00000376665.4 |
GATA1
|
GATA binding protein 1 |
| chr21_-_44261854 | 0.07 |
ENST00000431166.1
|
DNMT3L
|
DNA methyltransferase 3 like |
| chr5_-_142685654 | 0.07 |
ENST00000378046.5
ENST00000619447.4 |
FGF1
|
fibroblast growth factor 1 |
| chr6_+_18155399 | 0.06 |
ENST00000650836.2
ENST00000449850.2 ENST00000297792.9 |
KDM1B
|
lysine demethylase 1B |
| chr14_+_67533282 | 0.06 |
ENST00000329153.10
|
PLEKHH1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chr5_+_140175205 | 0.06 |
ENST00000644078.1
|
CYSTM1
|
cysteine rich transmembrane module containing 1 |
| chr17_+_80170012 | 0.06 |
ENST00000573882.5
ENST00000648509.2 ENST00000575465.6 ENST00000651672.1 |
CARD14
|
caspase recruitment domain family member 14 |
| chr7_+_118214633 | 0.06 |
ENST00000477532.5
|
ANKRD7
|
ankyrin repeat domain 7 |
| chr18_-_75208417 | 0.06 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr7_+_3301242 | 0.06 |
ENST00000404826.7
|
SDK1
|
sidekick cell adhesion molecule 1 |
| chr14_+_96204679 | 0.06 |
ENST00000542454.2
ENST00000539359.1 ENST00000554311.2 ENST00000553811.1 |
BDKRB2
ENSG00000258691.1
|
bradykinin receptor B2 novel protein |
| chr8_-_144416481 | 0.06 |
ENST00000276833.9
|
SLC39A4
|
solute carrier family 39 member 4 |
| chr11_-_72722302 | 0.06 |
ENST00000334211.12
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_+_218270392 | 0.06 |
ENST00000248451.7
ENST00000273077.9 |
PNKD
|
PNKD metallo-beta-lactamase domain containing |
| chr4_+_7192519 | 0.06 |
ENST00000507866.6
|
SORCS2
|
sortilin related VPS10 domain containing receptor 2 |
| chr5_-_176034680 | 0.06 |
ENST00000514861.5
|
THOC3
|
THO complex 3 |
| chr16_-_90008988 | 0.06 |
ENST00000568662.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr12_+_8509460 | 0.06 |
ENST00000382064.6
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chr2_+_102761963 | 0.06 |
ENST00000640575.2
ENST00000412401.3 |
TMEM182
|
transmembrane protein 182 |
| chr14_+_24161257 | 0.06 |
ENST00000396864.8
ENST00000557894.5 ENST00000559284.5 ENST00000560275.5 |
IRF9
|
interferon regulatory factor 9 |
| chr15_-_38564635 | 0.06 |
ENST00000450598.6
ENST00000559830.5 ENST00000558164.5 ENST00000539159.5 ENST00000310803.10 |
RASGRP1
|
RAS guanyl releasing protein 1 |
| chr11_-_65614195 | 0.06 |
ENST00000309100.8
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr20_-_32207708 | 0.06 |
ENST00000246229.5
|
PLAGL2
|
PLAG1 like zinc finger 2 |
| chr22_-_37007818 | 0.06 |
ENST00000405091.6
|
TEX33
|
testis expressed 33 |
| chr22_-_37007798 | 0.06 |
ENST00000402860.7
|
TEX33
|
testis expressed 33 |
| chr10_-_28303051 | 0.06 |
ENST00000683449.1
|
MPP7
|
membrane palmitoylated protein 7 |
| chr22_-_37007844 | 0.06 |
ENST00000381821.2
|
TEX33
|
testis expressed 33 |
| chr5_-_142686079 | 0.06 |
ENST00000337706.7
|
FGF1
|
fibroblast growth factor 1 |
| chr9_-_127873462 | 0.06 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr16_-_33845229 | 0.06 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chr11_+_124865425 | 0.05 |
ENST00000397801.6
|
ROBO3
|
roundabout guidance receptor 3 |
| chr12_-_48865863 | 0.05 |
ENST00000309739.6
|
RND1
|
Rho family GTPase 1 |
| chr9_+_132978651 | 0.05 |
ENST00000636137.1
|
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr7_-_44979003 | 0.05 |
ENST00000258787.12
ENST00000648014.1 |
MYO1G
|
myosin IG |
| chr12_-_57110284 | 0.05 |
ENST00000543873.6
ENST00000554663.5 ENST00000557635.5 |
STAT6
|
signal transducer and activator of transcription 6 |
| chr2_+_105241743 | 0.05 |
ENST00000258456.3
|
GPR45
|
G protein-coupled receptor 45 |
| chr16_-_68371005 | 0.05 |
ENST00000574662.1
|
SMPD3
|
sphingomyelin phosphodiesterase 3 |
| chr1_-_2194753 | 0.05 |
ENST00000378546.9
ENST00000400919.7 |
FAAP20
|
FA core complex associated protein 20 |
| chr15_+_44427793 | 0.05 |
ENST00000558966.5
ENST00000559793.5 ENST00000558968.1 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr19_+_17309531 | 0.05 |
ENST00000359866.9
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr2_-_89027700 | 0.05 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr12_+_54053815 | 0.05 |
ENST00000430889.3
|
HOXC4
|
homeobox C4 |
| chr17_-_1400168 | 0.05 |
ENST00000573026.1
ENST00000575977.1 ENST00000571732.5 ENST00000264335.13 |
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
| chr10_+_17809337 | 0.05 |
ENST00000569591.3
|
MRC1
|
mannose receptor C-type 1 |
| chr19_-_58499197 | 0.05 |
ENST00000594786.1
|
SLC27A5
|
solute carrier family 27 member 5 |
| chr1_-_44031352 | 0.05 |
ENST00000372306.7
ENST00000475075.6 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr14_-_106165730 | 0.05 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr3_+_194136138 | 0.05 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr18_-_43277482 | 0.05 |
ENST00000255224.8
ENST00000590752.5 ENST00000596867.1 |
SYT4
|
synaptotagmin 4 |
| chr19_+_45507470 | 0.05 |
ENST00000245932.11
ENST00000592139.1 ENST00000590603.1 |
VASP
|
vasodilator stimulated phosphoprotein |
| chr3_+_52779916 | 0.05 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr19_-_10517439 | 0.05 |
ENST00000333430.6
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr22_+_20967212 | 0.05 |
ENST00000434714.6
|
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr4_-_139302460 | 0.05 |
ENST00000394223.2
ENST00000676245.1 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr5_+_139648914 | 0.05 |
ENST00000502336.5
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr11_-_67353503 | 0.05 |
ENST00000539074.1
ENST00000530584.5 ENST00000531239.2 ENST00000312419.8 ENST00000529704.5 |
POLD4
|
DNA polymerase delta 4, accessory subunit |
| chr17_-_7290392 | 0.05 |
ENST00000571464.1
|
YBX2
|
Y-box binding protein 2 |
| chr3_-_195811857 | 0.05 |
ENST00000349607.8
ENST00000346145.8 |
MUC4
|
mucin 4, cell surface associated |
| chr17_+_1279655 | 0.05 |
ENST00000333813.4
|
TRARG1
|
trafficking regulator of GLUT4 (SLC2A4) 1 |
| chr22_-_37427433 | 0.05 |
ENST00000452946.1
ENST00000402918.7 |
ELFN2
ELFN2
|
extracellular leucine rich repeat and fibronectin type III domain containing 2 extracellular leucine rich repeat and fibronectin type III domain containing 2 |
| chr11_+_61481110 | 0.05 |
ENST00000338608.7
ENST00000432063.6 |
PPP1R32
|
protein phosphatase 1 regulatory subunit 32 |
| chr5_-_32312913 | 0.05 |
ENST00000280285.9
ENST00000382142.8 ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr5_-_151924846 | 0.05 |
ENST00000274576.9
|
GLRA1
|
glycine receptor alpha 1 |
| chr15_+_44427591 | 0.05 |
ENST00000558791.5
ENST00000260327.9 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr1_-_247331743 | 0.05 |
ENST00000294753.8
ENST00000682384.1 |
ZNF496
|
zinc finger protein 496 |
| chr17_-_82065525 | 0.05 |
ENST00000354321.11
|
DUS1L
|
dihydrouridine synthase 1 like |
| chr17_-_47957824 | 0.05 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chr5_-_151924824 | 0.05 |
ENST00000455880.2
|
GLRA1
|
glycine receptor alpha 1 |
| chr17_-_3557798 | 0.05 |
ENST00000301365.8
ENST00000572519.1 ENST00000576742.6 |
TRPV3
|
transient receptor potential cation channel subfamily V member 3 |
| chr18_-_75209126 | 0.05 |
ENST00000322342.4
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr17_-_49210567 | 0.04 |
ENST00000507680.6
|
GNGT2
|
G protein subunit gamma transducin 2 |
| chr3_-_149576203 | 0.04 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr6_+_54307856 | 0.04 |
ENST00000370869.7
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr3_-_195811916 | 0.04 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated |
| chr3_-_59049947 | 0.04 |
ENST00000491845.5
ENST00000472469.5 ENST00000482387.6 ENST00000295966.11 |
CFAP20DC
|
CFAP20 domain containing |
| chr9_+_132978687 | 0.04 |
ENST00000372122.4
ENST00000372123.5 |
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr19_-_37207074 | 0.04 |
ENST00000588873.1
|
ENSG00000267360.6
|
novel protein |
| chr17_+_77450737 | 0.04 |
ENST00000541152.6
ENST00000591704.5 |
SEPTIN9
|
septin 9 |
| chr1_+_153203424 | 0.04 |
ENST00000368747.2
|
LELP1
|
late cornified envelope like proline rich 1 |
| chr16_+_4788394 | 0.04 |
ENST00000615471.4
ENST00000589721.5 ENST00000615889.4 |
SMIM22
|
small integral membrane protein 22 |
| chr7_-_31340678 | 0.04 |
ENST00000297142.4
|
NEUROD6
|
neuronal differentiation 6 |
| chr19_-_3061403 | 0.04 |
ENST00000586839.1
|
TLE5
|
TLE family member 5, transcriptional modulator |
| chr7_-_27147774 | 0.04 |
ENST00000222728.3
|
HOXA6
|
homeobox A6 |
| chr16_+_57092570 | 0.04 |
ENST00000290776.13
ENST00000535318.6 |
CPNE2
|
copine 2 |
| chr3_+_23805941 | 0.04 |
ENST00000306627.8
ENST00000346855.7 |
UBE2E1
|
ubiquitin conjugating enzyme E2 E1 |
| chr10_+_11742361 | 0.04 |
ENST00000379215.9
ENST00000420401.5 |
ECHDC3
|
enoyl-CoA hydratase domain containing 3 |
| chr3_-_46882165 | 0.04 |
ENST00000431168.1
ENST00000654597.1 |
MYL3
|
myosin light chain 3 |
| chr1_-_6485433 | 0.04 |
ENST00000535355.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr16_+_4788411 | 0.04 |
ENST00000589327.5
|
SMIM22
|
small integral membrane protein 22 |
| chr16_+_2033264 | 0.04 |
ENST00000565855.5
ENST00000566198.1 |
SLC9A3R2
|
SLC9A3 regulator 2 |
| chrX_+_154420315 | 0.04 |
ENST00000651139.1
|
TAZ
|
tafazzin |
| chr15_-_55196608 | 0.04 |
ENST00000677989.1
ENST00000562895.2 ENST00000569386.2 |
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr11_+_129815841 | 0.04 |
ENST00000281441.8
|
TMEM45B
|
transmembrane protein 45B |
| chr1_-_154970735 | 0.04 |
ENST00000368445.9
ENST00000448116.7 ENST00000368449.8 |
SHC1
|
SHC adaptor protein 1 |
| chr8_+_22589240 | 0.04 |
ENST00000450780.6
ENST00000430850.6 ENST00000447849.2 ENST00000614502.4 ENST00000443561.3 |
ENSG00000248235.6
PDLIM2
|
novel protein PDZ and LIM domain 2 |
| chr4_+_105710809 | 0.04 |
ENST00000360505.9
ENST00000510865.5 ENST00000509336.5 |
GSTCD
|
glutathione S-transferase C-terminal domain containing |
| chr16_-_75495396 | 0.04 |
ENST00000332272.9
|
CHST6
|
carbohydrate sulfotransferase 6 |
| chr10_+_48306639 | 0.04 |
ENST00000395611.7
ENST00000432379.5 ENST00000374189.5 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr11_+_111255982 | 0.04 |
ENST00000637637.1
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr19_+_984314 | 0.04 |
ENST00000587001.6
ENST00000585809.6 ENST00000251289.9 ENST00000607440.5 |
WDR18
|
WD repeat domain 18 |
| chr1_+_15684284 | 0.04 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chr15_-_55196899 | 0.04 |
ENST00000677147.1
ENST00000260443.9 ENST00000677730.1 |
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr1_+_247416149 | 0.04 |
ENST00000366497.6
ENST00000391828.8 |
NLRP3
|
NLR family pyrin domain containing 3 |
| chr7_+_69967464 | 0.04 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr14_-_64823148 | 0.04 |
ENST00000389722.7
|
SPTB
|
spectrin beta, erythrocytic |
| chr1_+_247416166 | 0.04 |
ENST00000391827.3
ENST00000336119.8 |
NLRP3
|
NLR family pyrin domain containing 3 |
| chr3_+_173398438 | 0.04 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1 |
| chr14_-_65102468 | 0.04 |
ENST00000555932.5
ENST00000284165.10 ENST00000358402.8 ENST00000246163.2 ENST00000358664.9 ENST00000556979.5 ENST00000555667.5 ENST00000557746.5 ENST00000556443.5 ENST00000618858.4 ENST00000557277.5 ENST00000556892.5 |
MAX
|
MYC associated factor X |
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:2000974 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0032597 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.0 | GO:0072199 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.0 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.0 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.0 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |