Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
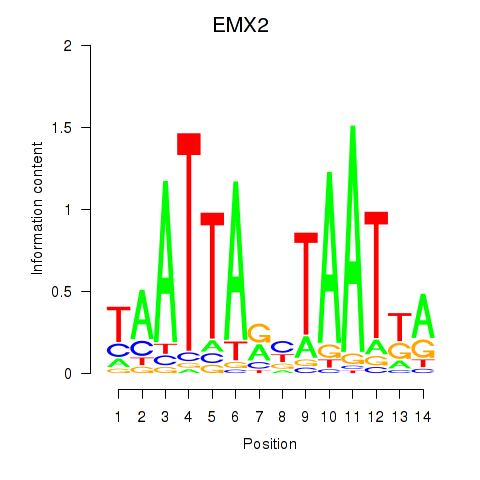
Results for EMX2
Z-value: 0.68
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.12 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg38_v1_chr10_+_117542416_117542445, hg38_v1_chr10_+_117543567_117543658, hg38_v1_chr10_+_117542721_117542754 | 0.26 | 5.4e-01 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91180365 | 0.87 |
ENST00000547937.5
|
DCN
|
decorin |
| chr6_+_72216442 | 0.86 |
ENST00000425662.6
ENST00000453976.6 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr11_-_63608542 | 0.80 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr10_-_91633057 | 0.57 |
ENST00000238994.6
|
PPP1R3C
|
protein phosphatase 1 regulatory subunit 3C |
| chr5_-_76623391 | 0.56 |
ENST00000296641.5
ENST00000504899.1 |
F2RL2
|
coagulation factor II thrombin receptor like 2 |
| chr9_-_76906041 | 0.52 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr5_-_111976925 | 0.49 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr4_+_70050431 | 0.45 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
| chr10_-_77638369 | 0.44 |
ENST00000372443.6
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr16_+_86566821 | 0.44 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr2_-_206218024 | 0.39 |
ENST00000407325.6
ENST00000612892.4 ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr8_-_13276491 | 0.39 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr10_-_77637902 | 0.39 |
ENST00000286627.10
ENST00000639486.1 ENST00000640523.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr4_-_137532452 | 0.37 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr10_-_20897288 | 0.34 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr10_-_77637721 | 0.33 |
ENST00000638848.1
ENST00000639406.1 ENST00000618048.2 ENST00000639120.1 ENST00000640834.1 ENST00000639601.1 ENST00000638514.1 ENST00000457953.6 ENST00000639090.1 ENST00000639489.1 ENST00000372440.6 ENST00000404771.8 ENST00000638203.1 ENST00000638306.1 ENST00000638351.1 ENST00000638606.1 ENST00000639591.1 ENST00000640182.1 ENST00000640605.1 ENST00000640141.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr10_-_77637789 | 0.33 |
ENST00000481070.1
ENST00000640969.1 ENST00000286628.14 ENST00000638991.1 ENST00000639913.1 ENST00000480683.2 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr19_+_49513353 | 0.32 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr6_+_72212802 | 0.32 |
ENST00000401910.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_189179754 | 0.31 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr2_-_98663464 | 0.31 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr6_+_72212887 | 0.28 |
ENST00000523963.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr17_+_1771688 | 0.28 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr1_+_77918128 | 0.28 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr6_+_101181254 | 0.27 |
ENST00000682090.1
ENST00000421544.6 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr12_+_59689337 | 0.25 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr6_+_135851681 | 0.25 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr5_+_141412979 | 0.24 |
ENST00000612503.1
ENST00000398610.3 |
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr6_-_29375291 | 0.23 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor family 12 subfamily D member 3 |
| chr7_+_90709530 | 0.22 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr4_-_88697810 | 0.22 |
ENST00000323061.7
|
NAP1L5
|
nucleosome assembly protein 1 like 5 |
| chr1_+_78649818 | 0.22 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr8_+_69492793 | 0.21 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr1_+_78004930 | 0.21 |
ENST00000370763.6
|
DNAJB4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr3_+_111674654 | 0.21 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr9_-_16728165 | 0.21 |
ENST00000603713.5
ENST00000603313.5 |
BNC2
|
basonuclin 2 |
| chr16_+_85027735 | 0.19 |
ENST00000258180.7
ENST00000538274.5 |
KIAA0513
|
KIAA0513 |
| chr10_+_15032193 | 0.19 |
ENST00000428897.5
ENST00000413672.5 |
OLAH
|
oleoyl-ACP hydrolase |
| chr4_+_55346213 | 0.18 |
ENST00000679836.1
ENST00000264228.9 ENST00000679707.1 |
SRD5A3
ENSG00000288695.1
|
steroid 5 alpha-reductase 3 novel protein, SRD5A3-RP11-177J6.1 readthrough |
| chr12_+_18242955 | 0.18 |
ENST00000676171.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr7_+_90709231 | 0.17 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr10_+_89332484 | 0.17 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr1_+_192809031 | 0.16 |
ENST00000235382.7
|
RGS2
|
regulator of G protein signaling 2 |
| chr21_-_30497160 | 0.15 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr7_+_138460238 | 0.15 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr17_-_69141878 | 0.15 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr11_+_66509079 | 0.15 |
ENST00000419755.3
|
ENSG00000256349.1
|
novel protein |
| chr1_+_177170916 | 0.15 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr1_-_60073750 | 0.14 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr2_+_201132958 | 0.14 |
ENST00000479953.6
ENST00000340870.6 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr1_+_152878312 | 0.14 |
ENST00000368765.4
|
SMCP
|
sperm mitochondria associated cysteine rich protein |
| chr2_+_201132928 | 0.14 |
ENST00000462763.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chrX_+_18425597 | 0.14 |
ENST00000623535.2
ENST00000674046.1 |
CDKL5
|
cyclin dependent kinase like 5 |
| chr14_+_34993240 | 0.14 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr11_-_124800630 | 0.13 |
ENST00000239614.8
ENST00000674284.1 |
MSANTD2
|
Myb/SANT DNA binding domain containing 2 |
| chr1_+_174875505 | 0.13 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr3_+_131026844 | 0.13 |
ENST00000510769.5
ENST00000383366.9 ENST00000510688.5 ENST00000511262.5 |
NEK11
|
NIMA related kinase 11 |
| chr9_-_96778053 | 0.13 |
ENST00000375231.5
ENST00000223428.9 |
ZNF510
|
zinc finger protein 510 |
| chr5_+_55160161 | 0.13 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr7_-_44848021 | 0.12 |
ENST00000349299.7
ENST00000521529.5 ENST00000350771.7 ENST00000222690.10 ENST00000381124.9 ENST00000437072.5 ENST00000446531.1 ENST00000308153.9 |
H2AZ2
|
H2A.Z variant histone 2 |
| chr5_-_132227472 | 0.12 |
ENST00000428369.6
|
P4HA2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr8_-_132675567 | 0.12 |
ENST00000519595.5
|
LRRC6
|
leucine rich repeat containing 6 |
| chr14_-_80231052 | 0.12 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chrX_+_55717733 | 0.12 |
ENST00000414239.5
ENST00000374941.9 |
RRAGB
|
Ras related GTP binding B |
| chr3_+_72888031 | 0.11 |
ENST00000389617.9
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr2_+_201132769 | 0.11 |
ENST00000494258.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr9_+_72577369 | 0.11 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr4_+_87832917 | 0.11 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr2_+_109794296 | 0.11 |
ENST00000430736.5
ENST00000016946.8 ENST00000441344.1 |
RGPD5
|
RANBP2 like and GRIP domain containing 5 |
| chr5_+_148394712 | 0.11 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chrX_+_55717796 | 0.11 |
ENST00000262850.7
|
RRAGB
|
Ras related GTP binding B |
| chr5_+_75611182 | 0.11 |
ENST00000672850.1
ENST00000672844.1 |
ANKDD1B
|
ankyrin repeat and death domain containing 1B |
| chr11_-_125111708 | 0.11 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr1_+_244835616 | 0.11 |
ENST00000366528.3
ENST00000411948.7 |
COX20
|
cytochrome c oxidase assembly factor COX20 |
| chr5_-_88823763 | 0.11 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr5_-_180861248 | 0.10 |
ENST00000502412.2
ENST00000512132.5 ENST00000506439.5 |
ZFP62
|
ZFP62 zinc finger protein |
| chr14_-_105242605 | 0.10 |
ENST00000549655.5
|
BRF1
|
BRF1 RNA polymerase III transcription initiation factor subunit |
| chr16_+_69565958 | 0.10 |
ENST00000349945.7
ENST00000354436.6 |
NFAT5
|
nuclear factor of activated T cells 5 |
| chr3_+_40477107 | 0.10 |
ENST00000314686.9
ENST00000447116.6 ENST00000429348.6 ENST00000432264.4 ENST00000456778.5 |
ZNF619
|
zinc finger protein 619 |
| chr4_+_155903688 | 0.10 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr17_-_36264514 | 0.10 |
ENST00000618620.4
ENST00000621034.1 |
TBC1D3I
|
TBC1 domain family member 3I |
| chr5_+_134967901 | 0.10 |
ENST00000282611.8
|
CATSPER3
|
cation channel sperm associated 3 |
| chr16_-_30091226 | 0.10 |
ENST00000279386.6
ENST00000627355.2 |
TBX6
|
T-box transcription factor 6 |
| chr3_-_49429304 | 0.10 |
ENST00000636166.1
ENST00000273598.8 ENST00000436744.2 |
ENSG00000283189.2
NICN1
|
novel protein nicolin 1 |
| chr10_-_122845850 | 0.10 |
ENST00000392790.6
|
CUZD1
|
CUB and zona pellucida like domains 1 |
| chrX_+_136648138 | 0.10 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr9_+_128566741 | 0.10 |
ENST00000630866.1
|
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr9_+_72577788 | 0.09 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr1_+_86547070 | 0.09 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr3_-_49429252 | 0.09 |
ENST00000615713.4
|
NICN1
|
nicolin 1 |
| chrX_+_136648214 | 0.09 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr3_-_127736329 | 0.09 |
ENST00000398101.7
|
MGLL
|
monoglyceride lipase |
| chr1_-_19923617 | 0.09 |
ENST00000375116.3
|
PLA2G2E
|
phospholipase A2 group IIE |
| chr4_-_88284616 | 0.09 |
ENST00000508256.5
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr18_+_34710307 | 0.09 |
ENST00000679796.1
|
DTNA
|
dystrobrevin alpha |
| chr4_-_88284590 | 0.09 |
ENST00000510548.6
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr3_-_155293665 | 0.08 |
ENST00000489090.2
|
STRIT1
|
small transmembrane regulator of ion transport 1 |
| chr6_-_26234978 | 0.08 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member |
| chr2_+_201132872 | 0.08 |
ENST00000470178.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr5_+_42548043 | 0.08 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr19_-_51417791 | 0.08 |
ENST00000353836.9
|
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr4_-_39032922 | 0.08 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr20_-_35411963 | 0.08 |
ENST00000349714.9
ENST00000438533.5 ENST00000359226.6 ENST00000374384.6 ENST00000374385.10 ENST00000424405.5 ENST00000397554.5 ENST00000374380.6 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr17_+_38127951 | 0.08 |
ENST00000621587.2
|
TBC1D3E
|
TBC1 domain family member 3E |
| chr17_+_38003976 | 0.08 |
ENST00000616101.4
|
TBC1D3D
|
TBC1 domain family member 3D |
| chr17_-_37989048 | 0.08 |
ENST00000617678.2
ENST00000612727.5 |
TBC1D3L
|
TBC1 domain family member 3L |
| chr12_-_11134644 | 0.08 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr6_+_28124596 | 0.08 |
ENST00000340487.5
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr17_-_15341577 | 0.08 |
ENST00000543896.1
ENST00000395930.6 ENST00000539245.5 ENST00000539316.1 |
TEKT3
|
tektin 3 |
| chr21_-_7825797 | 0.08 |
ENST00000617668.2
|
KCNE1B
|
potassium voltage-gated channel subfamily E regulatory subunit 1B |
| chr5_-_177780633 | 0.08 |
ENST00000513554.5
ENST00000440605.7 |
FAM153A
|
family with sequence similarity 153 member A |
| chr17_-_445939 | 0.08 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr14_+_61697622 | 0.08 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr1_+_224183197 | 0.08 |
ENST00000323699.9
|
DEGS1
|
delta 4-desaturase, sphingolipid 1 |
| chrX_+_77910656 | 0.07 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr15_+_64387828 | 0.07 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr19_-_34773184 | 0.07 |
ENST00000588760.1
ENST00000329285.13 ENST00000587354.6 |
ZNF599
|
zinc finger protein 599 |
| chr17_-_76141240 | 0.07 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr15_-_19988117 | 0.07 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr10_+_100462969 | 0.07 |
ENST00000343737.6
|
WNT8B
|
Wnt family member 8B |
| chr2_-_110577101 | 0.07 |
ENST00000330331.9
ENST00000446930.1 ENST00000329516.8 |
RGPD6
|
RANBP2 like and GRIP domain containing 6 |
| chr5_+_160009113 | 0.07 |
ENST00000522793.5
ENST00000682719.1 ENST00000684137.1 ENST00000683219.1 ENST00000684018.1 ENST00000231238.10 ENST00000682131.1 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr20_+_15196834 | 0.07 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr8_-_25424260 | 0.07 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1 |
| chr22_+_21665994 | 0.07 |
ENST00000680393.1
ENST00000679534.1 ENST00000679827.1 ENST00000681956.1 ENST00000681338.1 ENST00000680061.1 ENST00000679540.1 ENST00000679795.1 ENST00000335025.12 ENST00000398831.8 ENST00000679477.1 ENST00000626352.2 ENST00000458567.5 ENST00000680094.1 ENST00000680109.1 ENST00000406385.1 ENST00000680860.1 |
PPIL2
|
peptidylprolyl isomerase like 2 |
| chr3_-_186570308 | 0.07 |
ENST00000446782.5
|
TBCCD1
|
TBCC domain containing 1 |
| chr11_-_125111579 | 0.07 |
ENST00000532156.5
ENST00000532407.5 ENST00000279968.8 ENST00000527766.5 ENST00000529583.5 ENST00000524373.5 ENST00000527271.5 ENST00000526175.5 ENST00000529609.5 ENST00000682305.1 ENST00000533273.1 |
TMEM218
|
transmembrane protein 218 |
| chr8_-_132675533 | 0.07 |
ENST00000620350.5
ENST00000518642.5 ENST00000250173.5 |
LRRC6
|
leucine rich repeat containing 6 |
| chrX_+_47585212 | 0.07 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr11_+_117327829 | 0.07 |
ENST00000533153.5
ENST00000278935.8 ENST00000525416.5 |
CEP164
|
centrosomal protein 164 |
| chr2_-_112433519 | 0.07 |
ENST00000496537.1
ENST00000330575.9 ENST00000302558.8 |
RGPD8
|
RANBP2 like and GRIP domain containing 8 |
| chr2_-_206159410 | 0.06 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr1_+_84144260 | 0.06 |
ENST00000370685.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr7_+_144070313 | 0.06 |
ENST00000641441.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr5_+_143812161 | 0.06 |
ENST00000289448.4
|
HMHB1
|
histocompatibility minor HB-1 |
| chr19_-_58353482 | 0.06 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr10_-_95069489 | 0.06 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chr5_+_119476530 | 0.06 |
ENST00000645099.1
ENST00000513628.5 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr11_-_18236795 | 0.06 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr16_-_28363508 | 0.06 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family member B6 |
| chr2_+_161136901 | 0.06 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr7_-_150302980 | 0.06 |
ENST00000252071.8
|
ACTR3C
|
actin related protein 3C |
| chr4_-_100517991 | 0.06 |
ENST00000511970.5
ENST00000502569.1 ENST00000305864.7 ENST00000296420.9 |
EMCN
|
endomucin |
| chr4_+_26343156 | 0.06 |
ENST00000680928.1
ENST00000681260.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_+_36084079 | 0.06 |
ENST00000207457.8
|
TEKT2
|
tektin 2 |
| chr11_-_26572102 | 0.06 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr17_-_3298360 | 0.06 |
ENST00000323404.2
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1 |
| chr1_+_84181630 | 0.06 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr10_-_114144599 | 0.05 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr3_-_142000353 | 0.05 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr22_+_23856703 | 0.05 |
ENST00000345044.10
|
SLC2A11
|
solute carrier family 2 member 11 |
| chr12_+_55549602 | 0.05 |
ENST00000641569.1
ENST00000641851.1 |
OR6C4
|
olfactory receptor family 6 subfamily C member 4 |
| chr1_-_53940100 | 0.05 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr2_-_201643448 | 0.05 |
ENST00000409883.7
|
TMEM237
|
transmembrane protein 237 |
| chr11_-_26572254 | 0.05 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr12_+_26011713 | 0.05 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr2_+_11542662 | 0.05 |
ENST00000389825.7
ENST00000381483.6 |
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr13_+_108269629 | 0.05 |
ENST00000430559.5
ENST00000375887.9 |
TNFSF13B
|
TNF superfamily member 13b |
| chr9_-_21202205 | 0.05 |
ENST00000239347.3
|
IFNA7
|
interferon alpha 7 |
| chr6_+_27957241 | 0.05 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor family 2 subfamily B member 6 |
| chr3_-_131026726 | 0.05 |
ENST00000514044.5
ENST00000264992.8 |
ASTE1
|
asteroid homolog 1 |
| chr4_-_88284747 | 0.05 |
ENST00000514204.1
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr11_+_59511368 | 0.05 |
ENST00000641278.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chr16_+_28637654 | 0.05 |
ENST00000529716.5
|
NPIPB8
|
nuclear pore complex interacting protein family member B8 |
| chr9_-_21217311 | 0.05 |
ENST00000380216.1
|
IFNA16
|
interferon alpha 16 |
| chr1_-_248277976 | 0.05 |
ENST00000641220.1
|
OR2T33
|
olfactory receptor family 2 subfamily T member 33 |
| chr2_-_152175715 | 0.05 |
ENST00000263904.5
|
STAM2
|
signal transducing adaptor molecule 2 |
| chr3_-_47513677 | 0.05 |
ENST00000296149.9
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr11_-_26572130 | 0.05 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr1_+_197413827 | 0.05 |
ENST00000367397.1
ENST00000681519.1 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr22_-_40533808 | 0.04 |
ENST00000422851.1
ENST00000651694.1 ENST00000652095.2 |
MRTFA
|
myocardin related transcription factor A |
| chr4_+_94207596 | 0.04 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chrX_+_56729231 | 0.04 |
ENST00000637096.1
ENST00000374922.9 ENST00000423617.2 |
NBDY
|
negative regulator of P-body association |
| chr6_+_101398788 | 0.04 |
ENST00000369138.5
ENST00000413795.5 ENST00000358361.7 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr11_+_2384603 | 0.04 |
ENST00000527343.5
ENST00000464784.6 |
CD81
|
CD81 molecule |
| chr9_+_122371014 | 0.04 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr11_-_94973541 | 0.04 |
ENST00000279839.8
|
CWC15
|
CWC15 spliceosome associated protein homolog |
| chr4_-_112516176 | 0.04 |
ENST00000313341.4
|
NEUROG2
|
neurogenin 2 |
| chr12_+_8157034 | 0.04 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr14_-_106470788 | 0.04 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr1_-_159714581 | 0.04 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr6_-_75363003 | 0.04 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr22_+_31754862 | 0.04 |
ENST00000382111.6
ENST00000645407.1 ENST00000646701.1 |
DEPDC5
ENSG00000285404.1
|
DEP domain containing 5, GATOR1 subcomplex subunit novel protein, DEPDC5-YWHAH readthrough |
| chr17_-_41184895 | 0.04 |
ENST00000620667.1
ENST00000398472.2 |
KRTAP4-1
|
keratin associated protein 4-1 |
| chr5_+_129748091 | 0.04 |
ENST00000564719.2
|
MINAR2
|
membrane integral NOTCH2 associated receptor 2 |
| chr3_-_58537283 | 0.04 |
ENST00000459701.6
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr12_-_76083926 | 0.04 |
ENST00000551992.5
|
NAP1L1
|
nucleosome assembly protein 1 like 1 |
| chr10_-_73655984 | 0.04 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like |
| chr19_+_14583076 | 0.04 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr12_-_118359105 | 0.04 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr15_-_78621272 | 0.04 |
ENST00000348639.7
|
CHRNA3
|
cholinergic receptor nicotinic alpha 3 subunit |
| chr17_+_29941605 | 0.04 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chrX_+_91434829 | 0.04 |
ENST00000312600.4
|
PABPC5
|
poly(A) binding protein cytoplasmic 5 |
| chr3_+_159839847 | 0.04 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr3_-_58537181 | 0.04 |
ENST00000302819.10
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr17_-_4263847 | 0.04 |
ENST00000570535.5
ENST00000574367.5 ENST00000341657.9 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr2_+_206159884 | 0.04 |
ENST00000392222.7
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr16_+_56961942 | 0.04 |
ENST00000200676.8
ENST00000566128.1 |
CETP
|
cholesteryl ester transfer protein |
| chr19_-_51417700 | 0.04 |
ENST00000529627.1
ENST00000439889.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr9_-_14314132 | 0.04 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B |
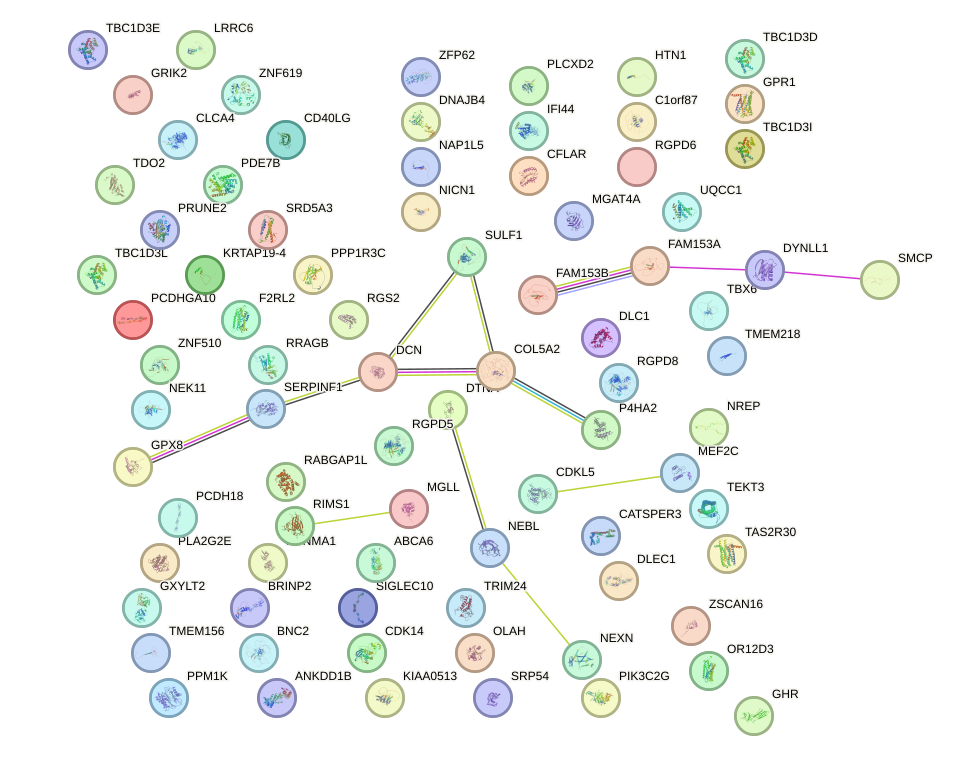
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 1.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.5 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0070244 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 1.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.2 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |