Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
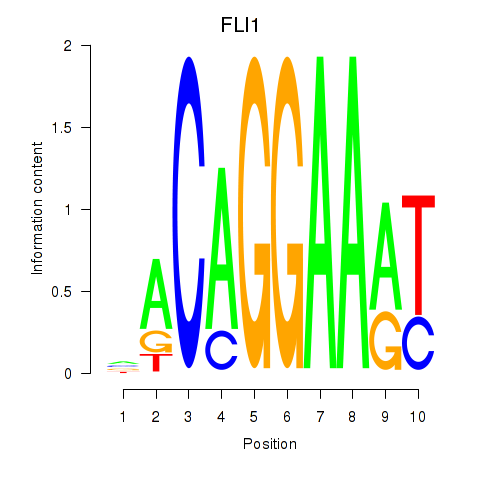
Results for FLI1
Z-value: 0.44
Transcription factors associated with FLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FLI1
|
ENSG00000151702.17 | Fli-1 proto-oncogene, ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FLI1 | hg38_v1_chr11_+_128694052_128694108, hg38_v1_chr11_+_128693887_128693931 | -0.68 | 6.4e-02 | Click! |
Activity profile of FLI1 motif
Sorted Z-values of FLI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_31321152 | 0.54 |
ENST00000446923.7
|
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr5_+_95731300 | 0.35 |
ENST00000379982.8
|
RHOBTB3
|
Rho related BTB domain containing 3 |
| chr7_+_100602344 | 0.34 |
ENST00000223061.6
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chr17_-_19387170 | 0.33 |
ENST00000395592.6
ENST00000299610.5 |
MFAP4
|
microfibril associated protein 4 |
| chr17_-_55732074 | 0.32 |
ENST00000575734.5
|
TMEM100
|
transmembrane protein 100 |
| chr1_+_183805105 | 0.28 |
ENST00000360851.4
|
RGL1
|
ral guanine nucleotide dissociation stimulator like 1 |
| chr2_-_187554351 | 0.27 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr2_-_224982420 | 0.27 |
ENST00000645028.1
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr2_-_224947030 | 0.26 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr5_+_157266079 | 0.26 |
ENST00000616178.4
ENST00000522463.5 ENST00000435847.6 ENST00000620254.5 ENST00000521420.5 ENST00000617629.4 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_153545793 | 0.26 |
ENST00000354332.8
ENST00000368716.9 |
S100A4
|
S100 calcium binding protein A4 |
| chr2_-_237414127 | 0.25 |
ENST00000472056.5
|
COL6A3
|
collagen type VI alpha 3 chain |
| chr2_-_237414157 | 0.24 |
ENST00000295550.9
ENST00000353578.9 ENST00000392004.7 ENST00000433762.1 ENST00000392003.6 |
COL6A3
|
collagen type VI alpha 3 chain |
| chr10_+_69088096 | 0.24 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr10_+_68106109 | 0.24 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr14_-_105940235 | 0.20 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr1_+_159204860 | 0.20 |
ENST00000368122.4
ENST00000368121.6 |
ACKR1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr1_+_154327737 | 0.19 |
ENST00000672630.1
|
ATP8B2
|
ATPase phospholipid transporting 8B2 |
| chr13_-_33185994 | 0.18 |
ENST00000255486.8
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr12_-_108339300 | 0.18 |
ENST00000550402.6
ENST00000552995.5 ENST00000312143.11 |
CMKLR1
|
chemerin chemokine-like receptor 1 |
| chr3_-_122022122 | 0.18 |
ENST00000393631.5
ENST00000273691.7 ENST00000344209.10 |
ILDR1
|
immunoglobulin like domain containing receptor 1 |
| chr4_+_54229261 | 0.18 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr5_-_76623391 | 0.18 |
ENST00000296641.5
ENST00000504899.1 |
F2RL2
|
coagulation factor II thrombin receptor like 2 |
| chr1_+_15305735 | 0.18 |
ENST00000375997.8
ENST00000524761.5 ENST00000375995.3 |
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr2_-_68319887 | 0.18 |
ENST00000409862.1
ENST00000263655.4 |
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr5_-_147401591 | 0.17 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr18_-_54959391 | 0.17 |
ENST00000591504.6
|
CCDC68
|
coiled-coil domain containing 68 |
| chr8_-_13514821 | 0.17 |
ENST00000276297.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr13_+_75788838 | 0.17 |
ENST00000497947.6
|
LMO7
|
LIM domain 7 |
| chr6_-_52577012 | 0.16 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr1_-_153544997 | 0.16 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr3_+_148697784 | 0.16 |
ENST00000497524.5
ENST00000418473.7 ENST00000349243.8 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor type 1 |
| chr21_+_42653734 | 0.16 |
ENST00000335512.8
ENST00000328862.10 ENST00000335440.10 ENST00000380328.6 ENST00000398225.7 ENST00000398227.7 ENST00000398229.7 ENST00000398232.7 ENST00000398234.7 ENST00000398236.7 ENST00000349112.7 ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr17_-_68601357 | 0.16 |
ENST00000592554.2
|
FAM20A
|
FAM20A golgi associated secretory pathway pseudokinase |
| chr4_-_107036302 | 0.16 |
ENST00000285311.8
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr9_+_87498491 | 0.16 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1 |
| chr11_-_82997394 | 0.15 |
ENST00000525117.5
ENST00000532548.5 |
RAB30
|
RAB30, member RAS oncogene family |
| chr5_+_68288346 | 0.15 |
ENST00000320694.12
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr2_-_174597795 | 0.15 |
ENST00000679041.1
|
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr1_-_167090370 | 0.15 |
ENST00000367868.4
|
GPA33
|
glycoprotein A33 |
| chr3_-_58577367 | 0.14 |
ENST00000464064.5
ENST00000360997.7 |
FAM107A
|
family with sequence similarity 107 member A |
| chr2_-_174634566 | 0.14 |
ENST00000392547.6
|
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr21_-_38121331 | 0.14 |
ENST00000482032.1
ENST00000398948.5 ENST00000328264.7 ENST00000645093.1 |
DSCR4
KCNJ6
|
Down syndrome critical region 4 potassium inwardly rectifying channel subfamily J member 6 |
| chr3_-_195583931 | 0.14 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr11_-_82997477 | 0.14 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr12_+_53046969 | 0.14 |
ENST00000379902.7
|
TNS2
|
tensin 2 |
| chr9_+_76459152 | 0.14 |
ENST00000444201.6
ENST00000376730.5 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1 |
| chr5_+_141359970 | 0.14 |
ENST00000522605.2
ENST00000622527.1 |
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr1_-_85578345 | 0.13 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr10_-_126388455 | 0.13 |
ENST00000368679.8
ENST00000368676.8 ENST00000448723.2 |
ADAM12
|
ADAM metallopeptidase domain 12 |
| chr14_+_100376398 | 0.13 |
ENST00000554998.5
ENST00000402312.8 ENST00000335290.10 ENST00000554175.5 |
WDR25
|
WD repeat domain 25 |
| chr5_+_150672057 | 0.13 |
ENST00000520112.1
|
MYOZ3
|
myozenin 3 |
| chr4_-_184825960 | 0.13 |
ENST00000281455.7
|
ACSL1
|
acyl-CoA synthetase long chain family member 1 |
| chr10_-_43574555 | 0.13 |
ENST00000374446.7
ENST00000535642.5 ENST00000426961.1 |
ZNF239
|
zinc finger protein 239 |
| chr10_-_79445617 | 0.13 |
ENST00000372336.4
|
ZCCHC24
|
zinc finger CCHC-type containing 24 |
| chr4_-_184826030 | 0.13 |
ENST00000507295.5
ENST00000504900.5 ENST00000454703.6 |
ACSL1
|
acyl-CoA synthetase long chain family member 1 |
| chr7_+_93921720 | 0.13 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr8_+_38820332 | 0.13 |
ENST00000518809.5
ENST00000520611.1 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr16_-_3305397 | 0.13 |
ENST00000396862.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr3_-_58577648 | 0.12 |
ENST00000394481.5
|
FAM107A
|
family with sequence similarity 107 member A |
| chr7_+_101127095 | 0.12 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1 |
| chr8_+_22565236 | 0.12 |
ENST00000523900.5
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr6_+_72212887 | 0.12 |
ENST00000523963.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_187554473 | 0.12 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr3_+_9902619 | 0.12 |
ENST00000421412.5
|
IL17RE
|
interleukin 17 receptor E |
| chr8_+_38728550 | 0.12 |
ENST00000520340.5
ENST00000518415.5 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr14_+_51860391 | 0.12 |
ENST00000335281.8
|
GNG2
|
G protein subunit gamma 2 |
| chr6_+_72212802 | 0.12 |
ENST00000401910.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_199458689 | 0.12 |
ENST00000443023.5
|
SATB2
|
SATB homeobox 2 |
| chr9_+_2017383 | 0.11 |
ENST00000382194.6
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_+_1056776 | 0.11 |
ENST00000650072.1
|
WDR37
|
WD repeat domain 37 |
| chr16_+_53208438 | 0.11 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_+_181223270 | 0.11 |
ENST00000315073.10
ENST00000351937.9 |
TRIM41
|
tripartite motif containing 41 |
| chr7_+_142855060 | 0.11 |
ENST00000619012.4
ENST00000652003.1 |
EPHB6
|
EPH receptor B6 |
| chr8_-_13514744 | 0.11 |
ENST00000316609.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr16_+_20806517 | 0.11 |
ENST00000348433.10
ENST00000568501.5 ENST00000261377.11 ENST00000566276.5 |
REXO5
|
RNA exonuclease 5 |
| chr2_-_197310646 | 0.11 |
ENST00000647377.1
|
ANKRD44
|
ankyrin repeat domain 44 |
| chr5_-_151686908 | 0.11 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr17_+_68512878 | 0.11 |
ENST00000585981.5
ENST00000589480.5 ENST00000585815.5 |
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr6_-_56851888 | 0.11 |
ENST00000312431.10
ENST00000520645.5 |
DST
|
dystonin |
| chr7_+_48171451 | 0.11 |
ENST00000435803.6
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr1_+_84144260 | 0.11 |
ENST00000370685.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_165294031 | 0.10 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr12_-_105236074 | 0.10 |
ENST00000551662.5
ENST00000553097.5 ENST00000258530.8 |
APPL2
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 |
| chr9_-_35732122 | 0.10 |
ENST00000314888.10
|
TLN1
|
talin 1 |
| chr5_-_36066874 | 0.10 |
ENST00000282507.8
ENST00000513300.5 |
UGT3A2
|
UDP glycosyltransferase family 3 member A2 |
| chr17_+_68512379 | 0.10 |
ENST00000392711.5
ENST00000585427.5 ENST00000589228.6 ENST00000536854.6 ENST00000588702.5 ENST00000589309.5 |
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr9_-_137028878 | 0.10 |
ENST00000625103.1
ENST00000614293.4 |
ABCA2
|
ATP binding cassette subfamily A member 2 |
| chr2_+_64989343 | 0.10 |
ENST00000234256.4
|
SLC1A4
|
solute carrier family 1 member 4 |
| chr2_+_65228122 | 0.10 |
ENST00000542850.2
|
ACTR2
|
actin related protein 2 |
| chrX_+_51803007 | 0.10 |
ENST00000375772.7
|
MAGED1
|
MAGE family member D1 |
| chr2_-_199457931 | 0.10 |
ENST00000417098.6
|
SATB2
|
SATB homeobox 2 |
| chr14_-_100376251 | 0.10 |
ENST00000556645.5
ENST00000556209.5 ENST00000556504.5 ENST00000556435.5 ENST00000554772.5 ENST00000553581.1 ENST00000553769.6 ENST00000554605.5 ENST00000557722.5 ENST00000553413.5 ENST00000553524.5 ENST00000358655.8 |
WARS1
|
tryptophanyl-tRNA synthetase 1 |
| chr3_+_48465811 | 0.10 |
ENST00000433541.1
ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr4_+_159241016 | 0.10 |
ENST00000644902.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr14_+_105486867 | 0.10 |
ENST00000409393.6
ENST00000392531.4 |
CRIP1
|
cysteine rich protein 1 |
| chr15_+_92900338 | 0.10 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr17_-_37542361 | 0.10 |
ENST00000614196.1
|
SYNRG
|
synergin gamma |
| chr9_+_18474100 | 0.10 |
ENST00000327883.11
ENST00000431052.6 ENST00000380570.8 ENST00000380548.9 |
ADAMTSL1
|
ADAMTS like 1 |
| chr16_+_20806698 | 0.10 |
ENST00000564274.5
ENST00000563068.1 |
REXO5
|
RNA exonuclease 5 |
| chr12_+_54028433 | 0.10 |
ENST00000243108.5
|
HOXC6
|
homeobox C6 |
| chr9_+_18474206 | 0.10 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS like 1 |
| chr16_+_1528674 | 0.10 |
ENST00000253934.9
|
TMEM204
|
transmembrane protein 204 |
| chr19_+_15107369 | 0.10 |
ENST00000342784.7
ENST00000597977.5 ENST00000600440.5 |
SYDE1
|
synapse defective Rho GTPase homolog 1 |
| chr19_+_56567461 | 0.10 |
ENST00000330619.13
|
ZNF470
|
zinc finger protein 470 |
| chr12_+_93677556 | 0.09 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr12_+_93677352 | 0.09 |
ENST00000552983.5
ENST00000332896.8 ENST00000552033.5 ENST00000548483.5 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr3_+_51942323 | 0.09 |
ENST00000431474.6
ENST00000417220.6 ENST00000398755.8 ENST00000471971.6 |
PARP3
|
poly(ADP-ribose) polymerase family member 3 |
| chr2_-_165203870 | 0.09 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr8_+_232416 | 0.09 |
ENST00000518414.5
ENST00000521270.5 ENST00000518320.6 ENST00000398612.3 |
ZNF596
|
zinc finger protein 596 |
| chrX_+_54808334 | 0.09 |
ENST00000218439.8
|
MAGED2
|
MAGE family member D2 |
| chr6_+_52420992 | 0.09 |
ENST00000636954.1
ENST00000636566.1 ENST00000638075.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr2_+_102104563 | 0.09 |
ENST00000409589.5
ENST00000409329.5 |
IL1R1
|
interleukin 1 receptor type 1 |
| chr6_-_129710145 | 0.09 |
ENST00000368149.3
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr19_-_49813151 | 0.09 |
ENST00000528094.5
ENST00000526575.1 |
FUZ
|
fuzzy planar cell polarity protein |
| chr12_-_10454485 | 0.09 |
ENST00000408006.7
ENST00000544822.2 ENST00000536188.5 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr2_+_201451711 | 0.09 |
ENST00000194530.8
ENST00000392249.6 |
STRADB
|
STE20 related adaptor beta |
| chr14_+_20891385 | 0.09 |
ENST00000304639.4
|
RNASE3
|
ribonuclease A family member 3 |
| chr2_-_201451446 | 0.09 |
ENST00000332624.8
ENST00000430254.1 |
TRAK2
|
trafficking kinesin protein 2 |
| chr21_+_43865200 | 0.09 |
ENST00000291572.13
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chrX_-_56995426 | 0.09 |
ENST00000640768.1
ENST00000638619.1 |
SPIN3
|
spindlin family member 3 |
| chr1_-_153550083 | 0.09 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr14_+_105314711 | 0.09 |
ENST00000447393.6
ENST00000547217.5 |
PACS2
|
phosphofurin acidic cluster sorting protein 2 |
| chr12_-_10453330 | 0.09 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr9_+_2621556 | 0.09 |
ENST00000680746.1
|
VLDLR
|
very low density lipoprotein receptor |
| chr2_+_108621260 | 0.09 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr9_+_137788781 | 0.09 |
ENST00000482340.5
|
EHMT1
|
euchromatic histone lysine methyltransferase 1 |
| chr6_+_131808011 | 0.09 |
ENST00000647893.1
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr3_+_29281049 | 0.09 |
ENST00000383767.7
|
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr8_+_232137 | 0.09 |
ENST00000521145.5
ENST00000320552.6 ENST00000308811.8 ENST00000640035.1 ENST00000522866.5 |
ZNF596
|
zinc finger protein 596 |
| chr11_-_2137277 | 0.09 |
ENST00000381392.5
ENST00000381395.5 ENST00000418738.2 |
IGF2
|
insulin like growth factor 2 |
| chr15_-_52652031 | 0.09 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr3_+_29280945 | 0.09 |
ENST00000434693.6
|
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chrX_+_54808359 | 0.09 |
ENST00000375058.5
ENST00000375060.5 |
MAGED2
|
MAGE family member D2 |
| chrX_-_56995508 | 0.08 |
ENST00000374919.6
ENST00000639007.1 ENST00000639583.1 ENST00000638289.1 ENST00000639525.1 ENST00000638386.1 |
SPIN3
|
spindlin family member 3 |
| chr2_-_152175715 | 0.08 |
ENST00000263904.5
|
STAM2
|
signal transducing adaptor molecule 2 |
| chr6_+_31586859 | 0.08 |
ENST00000433492.5
|
LST1
|
leukocyte specific transcript 1 |
| chr19_-_49813223 | 0.08 |
ENST00000533418.5
|
FUZ
|
fuzzy planar cell polarity protein |
| chr2_-_165204042 | 0.08 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr9_+_2621766 | 0.08 |
ENST00000382100.8
|
VLDLR
|
very low density lipoprotein receptor |
| chr11_-_417385 | 0.08 |
ENST00000332725.7
|
SIGIRR
|
single Ig and TIR domain containing |
| chr12_+_21526287 | 0.08 |
ENST00000256969.7
|
SPX
|
spexin hormone |
| chr8_+_38728186 | 0.08 |
ENST00000519416.5
ENST00000520615.5 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr11_+_35662739 | 0.08 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44 |
| chr2_-_127642131 | 0.08 |
ENST00000426981.5
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr5_+_138084015 | 0.08 |
ENST00000504809.5
ENST00000506684.6 ENST00000398754.1 |
WNT8A
|
Wnt family member 8A |
| chr12_+_112418889 | 0.08 |
ENST00000392597.5
|
PTPN11
|
protein tyrosine phosphatase non-receptor type 11 |
| chr14_+_88005128 | 0.08 |
ENST00000267549.5
|
GPR65
|
G protein-coupled receptor 65 |
| chr16_+_69566041 | 0.08 |
ENST00000567239.5
|
NFAT5
|
nuclear factor of activated T cells 5 |
| chr14_+_54567100 | 0.08 |
ENST00000554335.6
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr4_-_139084289 | 0.08 |
ENST00000510408.5
ENST00000379549.7 ENST00000358635.7 |
ELF2
|
E74 like ETS transcription factor 2 |
| chr5_-_124744513 | 0.08 |
ENST00000504926.5
|
ZNF608
|
zinc finger protein 608 |
| chr5_-_138875290 | 0.08 |
ENST00000521094.2
ENST00000274711.7 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr1_-_159923717 | 0.08 |
ENST00000368096.5
|
TAGLN2
|
transgelin 2 |
| chr5_+_68292562 | 0.08 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr12_-_15221394 | 0.08 |
ENST00000537647.5
ENST00000256953.6 ENST00000546331.5 |
RERG
|
RAS like estrogen regulated growth inhibitor |
| chr5_+_134905100 | 0.08 |
ENST00000512783.5
ENST00000254908.11 |
PCBD2
|
pterin-4 alpha-carbinolamine dehydratase 2 |
| chr17_-_8376658 | 0.08 |
ENST00000643221.1
ENST00000647210.1 ENST00000649935.1 ENST00000396267.3 |
KRBA2
|
KRAB-A domain containing 2 |
| chr11_-_417304 | 0.08 |
ENST00000397632.7
|
SIGIRR
|
single Ig and TIR domain containing |
| chr17_+_37375974 | 0.08 |
ENST00000615133.2
ENST00000611038.4 |
C17orf78
|
chromosome 17 open reading frame 78 |
| chr9_-_136095268 | 0.08 |
ENST00000277554.4
|
NACC2
|
NACC family member 2 |
| chr6_+_10555787 | 0.08 |
ENST00000316170.9
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr2_+_24076817 | 0.08 |
ENST00000613899.4
|
FAM228B
|
family with sequence similarity 228 member B |
| chrX_-_153446051 | 0.08 |
ENST00000370231.3
|
TREX2
|
three prime repair exonuclease 2 |
| chr12_+_119667859 | 0.08 |
ENST00000541640.5
|
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr2_+_157257687 | 0.08 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr11_+_72080595 | 0.08 |
ENST00000647530.1
ENST00000539271.6 ENST00000642510.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr16_+_30896606 | 0.07 |
ENST00000279804.3
ENST00000395019.3 |
CTF1
|
cardiotrophin 1 |
| chrX_+_86714623 | 0.07 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr2_+_188292771 | 0.07 |
ENST00000359135.7
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr3_+_119597874 | 0.07 |
ENST00000488919.5
ENST00000273371.9 ENST00000495992.5 |
PLA1A
|
phospholipase A1 member A |
| chr5_-_38595396 | 0.07 |
ENST00000263409.8
|
LIFR
|
LIF receptor subunit alpha |
| chrX_+_65588368 | 0.07 |
ENST00000609672.5
|
MSN
|
moesin |
| chr19_-_57578872 | 0.07 |
ENST00000196489.4
|
ZNF416
|
zinc finger protein 416 |
| chr3_+_29281552 | 0.07 |
ENST00000452462.5
ENST00000456853.1 |
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr19_-_10339610 | 0.07 |
ENST00000589261.5
ENST00000160262.10 ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr19_+_12163049 | 0.07 |
ENST00000425827.5
ENST00000439995.5 ENST00000652580.1 ENST00000343979.6 ENST00000418338.1 |
ZNF136
|
zinc finger protein 136 |
| chr5_+_103120264 | 0.07 |
ENST00000358359.8
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr20_-_47355657 | 0.07 |
ENST00000311275.11
|
ZMYND8
|
zinc finger MYND-type containing 8 |
| chr1_-_36323593 | 0.07 |
ENST00000490466.2
|
EVA1B
|
eva-1 homolog B |
| chr10_-_49539112 | 0.07 |
ENST00000355832.10
ENST00000447839.7 |
ERCC6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr10_-_49539015 | 0.07 |
ENST00000681659.1
ENST00000680107.1 |
ERCC6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr19_-_49813259 | 0.07 |
ENST00000313777.9
|
FUZ
|
fuzzy planar cell polarity protein |
| chr2_+_201116143 | 0.07 |
ENST00000443227.5
ENST00000309955.8 ENST00000341222.10 ENST00000341582.10 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr10_+_114821744 | 0.07 |
ENST00000369250.7
ENST00000369246.1 ENST00000369248.9 |
FAM160B1
|
family with sequence similarity 160 member B1 |
| chr4_+_932426 | 0.07 |
ENST00000514453.5
ENST00000515492.5 ENST00000509508.5 ENST00000264771.9 ENST00000515740.5 ENST00000508204.5 ENST00000510493.5 ENST00000514546.5 ENST00000622959.3 |
TMEM175
|
transmembrane protein 175 |
| chr20_+_36214373 | 0.07 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr2_+_230327218 | 0.07 |
ENST00000243810.10
ENST00000396563.8 |
SP140L
|
SP140 nuclear body protein like |
| chr19_-_45424364 | 0.07 |
ENST00000589165.5
|
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr1_+_23959109 | 0.07 |
ENST00000471915.5
|
PNRC2
|
proline rich nuclear receptor coactivator 2 |
| chr2_+_230327160 | 0.07 |
ENST00000444636.5
ENST00000415673.7 |
SP140L
|
SP140 nuclear body protein like |
| chr6_+_25962792 | 0.07 |
ENST00000357085.5
|
TRIM38
|
tripartite motif containing 38 |
| chr3_+_29281441 | 0.07 |
ENST00000273139.13
ENST00000383766.6 |
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr2_-_24360445 | 0.07 |
ENST00000443927.5
ENST00000406921.7 ENST00000412011.5 ENST00000355123.9 |
ITSN2
|
intersectin 2 |
| chr11_-_3165264 | 0.07 |
ENST00000389989.7
ENST00000263650.12 |
OSBPL5
|
oxysterol binding protein like 5 |
| chr8_-_144787275 | 0.07 |
ENST00000343459.8
ENST00000429371.7 ENST00000534445.1 |
ZNF34
|
zinc finger protein 34 |
| chr19_+_1908258 | 0.07 |
ENST00000411971.5
ENST00000588907.2 |
SCAMP4
|
secretory carrier membrane protein 4 |
| chr19_+_57614211 | 0.07 |
ENST00000600344.1
ENST00000396161.10 ENST00000600883.1 |
ZNF134
|
zinc finger protein 134 |
| chr4_-_76234509 | 0.07 |
ENST00000638295.1
|
SCARB2
|
scavenger receptor class B member 2 |
| chr11_+_72080803 | 0.07 |
ENST00000423494.6
ENST00000539587.6 ENST00000536917.2 ENST00000538478.5 ENST00000324866.11 ENST00000643715.1 ENST00000439209.5 |
LRTOMT
ENSG00000284922.2
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of FLI1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.2 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.2 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.0 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0044007 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.0 | 0.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0046061 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.0 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:2001025 | response to cyclosporin A(GO:1905237) positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:1902228 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.0 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.0 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.0 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0044714 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 0.0 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.0 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.0 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |