Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
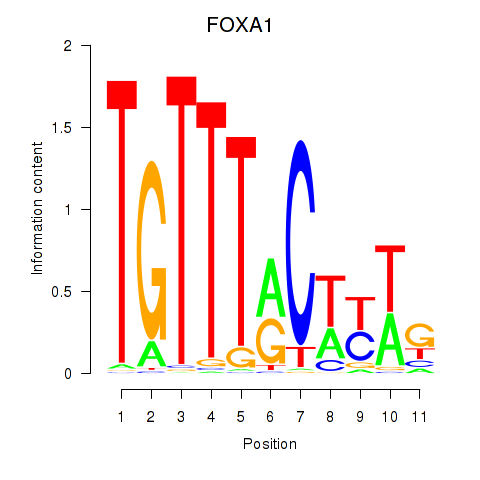
Results for FOXA1
Z-value: 1.21
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.8 | forkhead box A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg38_v1_chr14_-_37595224_37595281 | -0.26 | 5.3e-01 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91178520 | 1.41 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr9_-_92424427 | 1.21 |
ENST00000375550.5
|
OMD
|
osteomodulin |
| chr2_-_144516154 | 0.87 |
ENST00000637304.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_144516397 | 0.86 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_144431001 | 0.80 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_144430934 | 0.79 |
ENST00000638087.1
ENST00000638007.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_41510623 | 0.77 |
ENST00000328457.5
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3 |
| chr5_-_41510554 | 0.76 |
ENST00000377801.8
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3 |
| chr21_-_26843063 | 0.72 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843012 | 0.72 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr8_-_119673368 | 0.63 |
ENST00000427067.6
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr6_-_117425905 | 0.61 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr10_+_122560639 | 0.61 |
ENST00000344338.7
ENST00000330163.8 ENST00000652446.2 ENST00000666315.1 ENST00000368955.7 ENST00000368909.7 ENST00000368956.6 ENST00000619379.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr6_-_117425855 | 0.60 |
ENST00000368508.7
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr10_+_122560679 | 0.59 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr8_+_69492793 | 0.56 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr10_+_122560751 | 0.56 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chrX_+_16946862 | 0.56 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr18_-_63158208 | 0.52 |
ENST00000678301.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr2_-_213151590 | 0.49 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr13_+_75760362 | 0.46 |
ENST00000534657.5
|
LMO7
|
LIM domain 7 |
| chr1_+_222928415 | 0.45 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr6_-_87095059 | 0.44 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr6_-_56843153 | 0.42 |
ENST00000361203.7
ENST00000523817.1 |
DST
|
dystonin |
| chr17_-_19748355 | 0.39 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr5_-_128339191 | 0.38 |
ENST00000507835.5
|
FBN2
|
fibrillin 2 |
| chr9_-_20622479 | 0.37 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr2_+_172085499 | 0.37 |
ENST00000361725.5
ENST00000341900.6 |
DLX1
|
distal-less homeobox 1 |
| chr9_-_120477354 | 0.36 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr3_+_69936583 | 0.35 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr6_+_108656346 | 0.34 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr13_+_75760659 | 0.31 |
ENST00000526202.5
ENST00000465261.6 |
LMO7
|
LIM domain 7 |
| chr20_-_47355657 | 0.31 |
ENST00000311275.11
|
ZMYND8
|
zinc finger MYND-type containing 8 |
| chr5_+_150778733 | 0.31 |
ENST00000526627.2
|
SMIM3
|
small integral membrane protein 3 |
| chr3_+_141387801 | 0.30 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr9_-_86947496 | 0.29 |
ENST00000298743.9
|
GAS1
|
growth arrest specific 1 |
| chr2_+_168901290 | 0.29 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr1_-_177970213 | 0.28 |
ENST00000464631.6
|
SEC16B
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr3_-_146161167 | 0.28 |
ENST00000360060.7
ENST00000282903.10 |
PLOD2
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 |
| chr13_+_75760431 | 0.27 |
ENST00000321797.12
|
LMO7
|
LIM domain 7 |
| chr8_+_94895813 | 0.26 |
ENST00000396113.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr3_+_148827800 | 0.26 |
ENST00000282957.9
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 |
| chr1_+_40247926 | 0.25 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr5_+_102865805 | 0.25 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_148394712 | 0.25 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr2_-_233013228 | 0.24 |
ENST00000264051.8
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr15_+_96325935 | 0.24 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chrX_+_86714623 | 0.24 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr4_+_54229261 | 0.24 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr22_+_35066136 | 0.23 |
ENST00000308700.6
ENST00000404699.7 |
ISX
|
intestine specific homeobox |
| chr6_+_52420992 | 0.23 |
ENST00000636954.1
ENST00000636566.1 ENST00000638075.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr1_-_26913964 | 0.22 |
ENST00000254227.4
|
NR0B2
|
nuclear receptor subfamily 0 group B member 2 |
| chr5_-_138338325 | 0.22 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr8_+_76681208 | 0.22 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr12_-_71157872 | 0.21 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr12_-_71157992 | 0.21 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8 |
| chr20_+_33235987 | 0.21 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr10_+_102776237 | 0.21 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like |
| chr12_+_103587266 | 0.21 |
ENST00000388887.7
|
STAB2
|
stabilin 2 |
| chr9_+_6413191 | 0.20 |
ENST00000276893.10
|
UHRF2
|
ubiquitin like with PHD and ring finger domains 2 |
| chr5_+_98769273 | 0.20 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr16_-_87936529 | 0.19 |
ENST00000649794.3
ENST00000649158.1 ENST00000648177.1 |
CA5A
|
carbonic anhydrase 5A |
| chr18_+_75210789 | 0.19 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr3_-_71583713 | 0.19 |
ENST00000649528.3
ENST00000471386.3 ENST00000493089.7 |
FOXP1
|
forkhead box P1 |
| chr1_+_61082398 | 0.18 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A |
| chrX_+_136648138 | 0.18 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr2_+_87748087 | 0.18 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr18_+_3449413 | 0.17 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr2_+_151410090 | 0.17 |
ENST00000430328.6
|
RIF1
|
replication timing regulatory factor 1 |
| chr1_+_61081728 | 0.17 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr7_+_134891566 | 0.17 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chrX_+_22032301 | 0.17 |
ENST00000379374.5
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chrX_+_136648214 | 0.17 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr14_-_94323324 | 0.17 |
ENST00000341584.4
|
SERPINA6
|
serpin family A member 6 |
| chr7_+_134745460 | 0.17 |
ENST00000436461.6
|
CALD1
|
caldesmon 1 |
| chr2_+_203936755 | 0.17 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr19_+_38390055 | 0.16 |
ENST00000587947.5
ENST00000338502.8 |
SPRED3
|
sprouty related EVH1 domain containing 3 |
| chr20_+_33007695 | 0.16 |
ENST00000170150.4
|
BPIFB2
|
BPI fold containing family B member 2 |
| chr3_-_71583683 | 0.16 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr12_+_77830886 | 0.15 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chr5_+_96662969 | 0.15 |
ENST00000514845.5
ENST00000675663.1 |
CAST
|
calpastatin |
| chr20_-_44521989 | 0.15 |
ENST00000342374.5
ENST00000255175.5 |
SERINC3
|
serine incorporator 3 |
| chr4_+_164754116 | 0.15 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr15_+_75043263 | 0.14 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr16_-_4846196 | 0.14 |
ENST00000589389.5
|
GLYR1
|
glyoxylate reductase 1 homolog |
| chr10_-_79560386 | 0.14 |
ENST00000372327.9
ENST00000417041.1 ENST00000640627.1 ENST00000372325.7 |
SFTPA2
|
surfactant protein A2 |
| chr7_+_135148041 | 0.14 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr11_-_9265302 | 0.14 |
ENST00000328194.8
|
DENND5A
|
DENN domain containing 5A |
| chr13_+_50015438 | 0.14 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr8_+_94895837 | 0.14 |
ENST00000519136.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr1_+_116111395 | 0.14 |
ENST00000684484.1
ENST00000369500.4 |
MAB21L3
|
mab-21 like 3 |
| chr13_+_50015254 | 0.14 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr6_+_73696145 | 0.13 |
ENST00000287097.6
|
CD109
|
CD109 molecule |
| chr6_+_73695779 | 0.13 |
ENST00000422508.6
ENST00000437994.6 |
CD109
|
CD109 molecule |
| chr16_+_4846652 | 0.13 |
ENST00000592120.5
|
UBN1
|
ubinuclein 1 |
| chr12_-_55729660 | 0.13 |
ENST00000546457.1
ENST00000549117.5 |
CD63
|
CD63 molecule |
| chr12_+_96194365 | 0.13 |
ENST00000228741.8
ENST00000547249.1 |
ELK3
|
ETS transcription factor ELK3 |
| chr1_+_61082553 | 0.13 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr13_-_45201027 | 0.13 |
ENST00000379108.2
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr7_+_134891400 | 0.12 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr5_-_134174765 | 0.12 |
ENST00000520417.1
|
SKP1
|
S-phase kinase associated protein 1 |
| chr12_-_48004496 | 0.12 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain |
| chr14_+_64504574 | 0.12 |
ENST00000358738.3
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_+_121102666 | 0.12 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr7_-_93226449 | 0.12 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr11_-_116792386 | 0.12 |
ENST00000433069.2
ENST00000542499.5 |
APOA5
|
apolipoprotein A5 |
| chr8_+_94895763 | 0.11 |
ENST00000523378.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr12_+_96194501 | 0.11 |
ENST00000552142.5
|
ELK3
|
ETS transcription factor ELK3 |
| chr21_-_42315336 | 0.11 |
ENST00000398431.2
ENST00000518498.3 |
TFF3
|
trefoil factor 3 |
| chr18_+_3449620 | 0.11 |
ENST00000405385.7
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr10_+_113709261 | 0.11 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chrX_-_48003949 | 0.11 |
ENST00000396965.5
ENST00000376943.8 |
ZNF182
|
zinc finger protein 182 |
| chr5_+_96663010 | 0.10 |
ENST00000506811.5
ENST00000514055.5 ENST00000508608.6 |
CAST
|
calpastatin |
| chr12_-_120327762 | 0.10 |
ENST00000308366.9
ENST00000423423.3 |
PLA2G1B
|
phospholipase A2 group IB |
| chr3_-_149792547 | 0.10 |
ENST00000446160.7
ENST00000462519.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr1_-_56966133 | 0.10 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr18_+_75210755 | 0.10 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr4_-_154612635 | 0.10 |
ENST00000407946.5
ENST00000405164.5 ENST00000336098.8 ENST00000393846.6 ENST00000404648.7 ENST00000443553.5 |
FGG
|
fibrinogen gamma chain |
| chr22_-_17219571 | 0.09 |
ENST00000610390.4
|
ADA2
|
adenosine deaminase 2 |
| chr2_-_55334529 | 0.09 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr20_+_45791930 | 0.09 |
ENST00000372622.8
|
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1 |
| chr11_-_102530738 | 0.09 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr19_+_49877660 | 0.09 |
ENST00000535102.6
|
TBC1D17
|
TBC1 domain family member 17 |
| chr14_+_64503943 | 0.09 |
ENST00000556965.1
ENST00000554015.5 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr22_-_40533808 | 0.09 |
ENST00000422851.1
ENST00000651694.1 ENST00000652095.2 |
MRTFA
|
myocardin related transcription factor A |
| chr10_+_89332484 | 0.09 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr19_+_49877694 | 0.09 |
ENST00000221543.10
|
TBC1D17
|
TBC1 domain family member 17 |
| chr11_-_124752247 | 0.09 |
ENST00000326621.10
|
VSIG2
|
V-set and immunoglobulin domain containing 2 |
| chr19_-_48390847 | 0.09 |
ENST00000597017.5
|
KDELR1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chrX_+_130339941 | 0.09 |
ENST00000218197.9
|
SLC25A14
|
solute carrier family 25 member 14 |
| chr1_-_56966006 | 0.09 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr1_-_152360004 | 0.08 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr1_+_43389889 | 0.08 |
ENST00000562955.2
ENST00000634258.3 |
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr2_-_87021844 | 0.08 |
ENST00000355705.4
ENST00000409310.6 |
PLGLB1
|
plasminogen like B1 |
| chr11_-_26722051 | 0.08 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr12_+_120978537 | 0.08 |
ENST00000257555.11
ENST00000400024.6 |
HNF1A
|
HNF1 homeobox A |
| chr19_+_42302098 | 0.08 |
ENST00000598490.1
ENST00000341747.8 |
PRR19
|
proline rich 19 |
| chr11_-_31812171 | 0.08 |
ENST00000639409.1
ENST00000640975.1 |
PAX6
|
paired box 6 |
| chr10_-_31031911 | 0.08 |
ENST00000375311.1
ENST00000413025.5 ENST00000436087.6 ENST00000452305.5 ENST00000442986.5 |
ZNF438
|
zinc finger protein 438 |
| chr22_-_17219424 | 0.08 |
ENST00000649540.1
ENST00000399837.8 ENST00000543038.1 |
ADA2
|
adenosine deaminase 2 |
| chr3_-_52231190 | 0.08 |
ENST00000494383.1
|
ENSG00000173366.11
|
novel twinfilin, actin-binding protein, homolog 2 (Drosophila) (TWF2) and toll-like receptor 9 (TLR9) protein |
| chr1_-_244860376 | 0.08 |
ENST00000638716.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U |
| chr1_+_200027605 | 0.08 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr17_-_41315706 | 0.08 |
ENST00000334202.5
|
KRTAP17-1
|
keratin associated protein 17-1 |
| chr4_+_74308463 | 0.07 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr6_-_89315291 | 0.07 |
ENST00000402938.4
|
GABRR2
|
gamma-aminobutyric acid type A receptor subunit rho2 |
| chr3_+_108602776 | 0.07 |
ENST00000497905.5
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chrX_+_38006551 | 0.07 |
ENST00000297875.7
|
SYTL5
|
synaptotagmin like 5 |
| chr17_-_42745025 | 0.07 |
ENST00000592492.5
ENST00000585893.5 ENST00000593214.5 ENST00000590078.5 ENST00000428826.7 ENST00000586382.5 ENST00000415827.6 ENST00000592743.5 ENST00000586089.5 |
EZH1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr11_+_35662739 | 0.07 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44 |
| chr4_+_169660062 | 0.07 |
ENST00000507875.5
ENST00000613795.4 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr16_-_49664225 | 0.07 |
ENST00000535559.5
|
ZNF423
|
zinc finger protein 423 |
| chr1_-_120100688 | 0.07 |
ENST00000652264.1
|
NOTCH2
|
notch receptor 2 |
| chrX_+_130339886 | 0.07 |
ENST00000543953.5
ENST00000612248.4 ENST00000424447.5 ENST00000545805.6 |
SLC25A14
|
solute carrier family 25 member 14 |
| chr15_+_71096941 | 0.07 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr2_+_102473219 | 0.07 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr2_+_151409878 | 0.07 |
ENST00000453091.6
ENST00000428287.6 ENST00000444746.7 ENST00000243326.9 ENST00000414861.6 |
RIF1
|
replication timing regulatory factor 1 |
| chr7_+_77840122 | 0.07 |
ENST00000450574.5
ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr2_+_65056382 | 0.07 |
ENST00000377990.7
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68 |
| chr1_+_74235377 | 0.07 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr3_+_138010143 | 0.07 |
ENST00000183605.10
|
CLDN18
|
claudin 18 |
| chr14_+_55661272 | 0.07 |
ENST00000555573.5
|
KTN1
|
kinectin 1 |
| chr20_+_33168148 | 0.07 |
ENST00000354932.6
|
BPIFA2
|
BPI fold containing family A member 2 |
| chr3_+_69936629 | 0.06 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr7_-_100100716 | 0.06 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr12_+_120978686 | 0.06 |
ENST00000541395.5
ENST00000544413.2 |
HNF1A
|
HNF1 homeobox A |
| chr3_+_137998735 | 0.06 |
ENST00000343735.8
|
CLDN18
|
claudin 18 |
| chr4_+_55853639 | 0.06 |
ENST00000381295.7
ENST00000346134.11 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr1_-_216423396 | 0.06 |
ENST00000366942.3
ENST00000674083.1 ENST00000307340.8 |
USH2A
|
usherin |
| chr6_-_89217339 | 0.06 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr5_-_149379286 | 0.06 |
ENST00000261796.4
|
IL17B
|
interleukin 17B |
| chr2_-_171160833 | 0.06 |
ENST00000360843.7
ENST00000431350.7 |
TLK1
|
tousled like kinase 1 |
| chr4_+_67558719 | 0.06 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr19_-_18791297 | 0.06 |
ENST00000542601.6
ENST00000222271.7 ENST00000425807.1 |
COMP
|
cartilage oligomeric matrix protein |
| chr19_+_49877425 | 0.06 |
ENST00000622860.4
|
TBC1D17
|
TBC1 domain family member 17 |
| chr4_-_110198579 | 0.06 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr11_+_844406 | 0.05 |
ENST00000397404.5
|
TSPAN4
|
tetraspanin 4 |
| chr10_+_18400562 | 0.05 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr4_-_110198650 | 0.05 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr5_+_96936071 | 0.05 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase |
| chr1_+_43389874 | 0.05 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr4_+_186266183 | 0.05 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr5_-_40755885 | 0.05 |
ENST00000636863.1
ENST00000637375.1 ENST00000337702.5 ENST00000636106.1 |
TTC33
|
tetratricopeptide repeat domain 33 |
| chr4_-_163613505 | 0.05 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr7_+_117014881 | 0.05 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_94925759 | 0.05 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr11_-_6440980 | 0.05 |
ENST00000265983.8
ENST00000615166.1 |
HPX
|
hemopexin |
| chr7_+_106865263 | 0.05 |
ENST00000440650.6
ENST00000496166.6 ENST00000473541.5 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr13_-_99258366 | 0.05 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr14_+_64504743 | 0.05 |
ENST00000683701.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr6_-_42048648 | 0.05 |
ENST00000502771.1
ENST00000508143.5 ENST00000514588.1 ENST00000510503.5 |
CCND3
|
cyclin D3 |
| chr22_-_21227637 | 0.05 |
ENST00000401924.5
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr3_-_45915698 | 0.04 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr20_-_54070520 | 0.04 |
ENST00000371435.6
ENST00000395961.7 |
BCAS1
|
brain enriched myelin associated protein 1 |
| chr9_-_14910421 | 0.04 |
ENST00000380880.4
|
FREM1
|
FRAS1 related extracellular matrix 1 |
| chr11_+_1157946 | 0.04 |
ENST00000621226.2
|
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming |
| chr1_-_43389768 | 0.04 |
ENST00000372455.4
ENST00000372457.9 ENST00000290663.10 |
MED8
|
mediator complex subunit 8 |
| chr12_-_68253502 | 0.04 |
ENST00000328087.6
ENST00000538666.6 |
IL22
|
interleukin 22 |
| chr1_-_66801276 | 0.04 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr3_+_148730100 | 0.04 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 1.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.5 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.5 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.4 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 1.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.3 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.0 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.0 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.0 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.0 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.0 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.4 | GO:0097431 | pericentriolar material(GO:0000242) mitotic spindle pole(GO:0097431) |
| 0.0 | 1.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0032302 | MutSalpha complex(GO:0032301) MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 0.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 3.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 1.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0032143 | double-strand/single-strand DNA junction binding(GO:0000406) single thymine insertion binding(GO:0032143) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |