Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
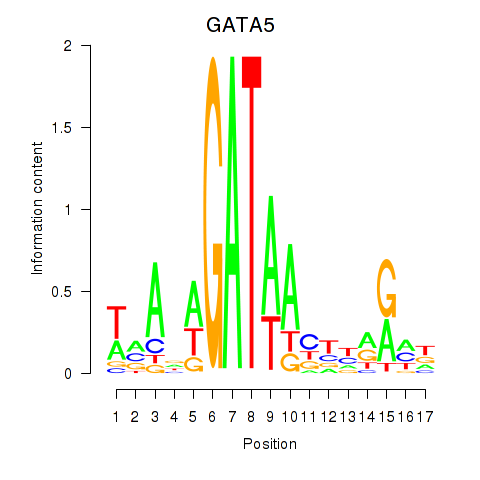
Results for GATA5
Z-value: 0.26
Transcription factors associated with GATA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA5
|
ENSG00000130700.7 | GATA binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA5 | hg38_v1_chr20_-_62475983_62476001 | 0.23 | 5.9e-01 | Click! |
Activity profile of GATA5 motif
Sorted Z-values of GATA5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_91946989 | 0.23 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr13_+_101489940 | 0.22 |
ENST00000376162.7
|
ITGBL1
|
integrin subunit beta like 1 |
| chr4_+_73740541 | 0.15 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr6_+_26500296 | 0.15 |
ENST00000684113.1
|
BTN1A1
|
butyrophilin subfamily 1 member A1 |
| chr5_+_93583212 | 0.14 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr8_-_23854796 | 0.11 |
ENST00000290271.7
|
STC1
|
stanniocalcin 1 |
| chr11_-_27700447 | 0.11 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor |
| chr1_+_78649818 | 0.10 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr11_-_27700472 | 0.10 |
ENST00000418212.5
ENST00000533246.5 |
BDNF
|
brain derived neurotrophic factor |
| chr9_-_92482350 | 0.09 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr9_-_120477354 | 0.09 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr8_+_27771942 | 0.09 |
ENST00000523566.5
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr6_+_85449584 | 0.09 |
ENST00000369651.7
|
NT5E
|
5'-nucleotidase ecto |
| chr1_+_21570303 | 0.08 |
ENST00000374830.2
|
ALPL
|
alkaline phosphatase, biomineralization associated |
| chr12_+_53097656 | 0.08 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr1_+_174875505 | 0.08 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr3_+_141426108 | 0.08 |
ENST00000441582.2
ENST00000510726.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_-_61001430 | 0.08 |
ENST00000357917.4
|
RHOBTB1
|
Rho related BTB domain containing 1 |
| chrM_+_12329 | 0.08 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr9_-_112175185 | 0.07 |
ENST00000355396.7
|
SUSD1
|
sushi domain containing 1 |
| chr17_+_64449037 | 0.07 |
ENST00000615220.4
ENST00000619286.5 ENST00000612535.4 ENST00000616498.4 |
MILR1
|
mast cell immunoglobulin like receptor 1 |
| chr11_+_124954108 | 0.07 |
ENST00000529051.5
|
CCDC15
|
coiled-coil domain containing 15 |
| chr13_+_108218366 | 0.07 |
ENST00000375898.4
|
ABHD13
|
abhydrolase domain containing 13 |
| chr3_-_179251615 | 0.07 |
ENST00000314235.9
ENST00000392685.6 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr1_+_224183197 | 0.06 |
ENST00000323699.9
|
DEGS1
|
delta 4-desaturase, sphingolipid 1 |
| chr7_+_142760398 | 0.06 |
ENST00000632998.1
|
PRSS2
|
serine protease 2 |
| chr3_-_138132381 | 0.06 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr19_-_44304968 | 0.06 |
ENST00000591609.1
ENST00000589799.5 ENST00000291182.9 ENST00000650576.1 ENST00000589248.5 |
ZNF235
|
zinc finger protein 235 |
| chr15_-_53733103 | 0.06 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr12_-_44876294 | 0.06 |
ENST00000429094.7
ENST00000551601.5 ENST00000549027.5 ENST00000452445.6 |
NELL2
|
neural EGFL like 2 |
| chr7_+_134646845 | 0.06 |
ENST00000344924.8
|
BPGM
|
bisphosphoglycerate mutase |
| chr15_+_99565950 | 0.06 |
ENST00000557785.5
ENST00000558049.5 ENST00000449277.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr7_+_116953238 | 0.05 |
ENST00000393446.6
|
ST7
|
suppression of tumorigenicity 7 |
| chr15_-_55408245 | 0.05 |
ENST00000563171.5
ENST00000425574.7 ENST00000442196.8 ENST00000564092.1 |
CCPG1
|
cell cycle progression 1 |
| chr7_+_134646798 | 0.05 |
ENST00000418040.5
ENST00000393132.2 |
BPGM
|
bisphosphoglycerate mutase |
| chr15_+_99565921 | 0.05 |
ENST00000558812.5
ENST00000338042.10 |
MEF2A
|
myocyte enhancer factor 2A |
| chr15_+_99566066 | 0.05 |
ENST00000557942.5
|
MEF2A
|
myocyte enhancer factor 2A |
| chr12_-_44875647 | 0.05 |
ENST00000395487.6
|
NELL2
|
neural EGFL like 2 |
| chr6_-_154356735 | 0.05 |
ENST00000367220.8
ENST00000265198.8 ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chrX_-_66033664 | 0.05 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_+_171485520 | 0.05 |
ENST00000647382.2
ENST00000392078.7 ENST00000367742.7 ENST00000338920.8 |
PRRC2C
|
proline rich coiled-coil 2C |
| chr9_-_112175264 | 0.05 |
ENST00000374264.6
|
SUSD1
|
sushi domain containing 1 |
| chr2_-_217842154 | 0.05 |
ENST00000446688.5
|
TNS1
|
tensin 1 |
| chr7_-_93148345 | 0.05 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr18_+_34709356 | 0.05 |
ENST00000585446.1
ENST00000681241.1 |
DTNA
|
dystrobrevin alpha |
| chr5_+_141223332 | 0.05 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chr2_+_201132958 | 0.05 |
ENST00000479953.6
ENST00000340870.6 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr17_+_20073538 | 0.04 |
ENST00000681116.1
ENST00000680572.1 ENST00000680604.1 ENST00000681875.1 ENST00000679058.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_-_158686700 | 0.04 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1 |
| chr10_-_91909476 | 0.04 |
ENST00000311575.6
|
FGFBP3
|
fibroblast growth factor binding protein 3 |
| chr3_+_156142962 | 0.04 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr9_-_124500986 | 0.04 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr2_-_46916020 | 0.04 |
ENST00000409800.5
ENST00000409218.5 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr10_+_15043937 | 0.04 |
ENST00000378228.8
ENST00000378217.3 |
OLAH
|
oleoyl-ACP hydrolase |
| chr12_-_130839230 | 0.04 |
ENST00000392373.7
ENST00000261653.10 |
STX2
|
syntaxin 2 |
| chr1_+_207104226 | 0.04 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha |
| chrX_+_30243715 | 0.04 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr17_-_15598797 | 0.04 |
ENST00000354433.7
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr8_+_93794816 | 0.04 |
ENST00000519845.5
ENST00000684343.1 |
TMEM67
|
transmembrane protein 67 |
| chr2_+_201132928 | 0.04 |
ENST00000462763.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr1_+_115029823 | 0.04 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr8_-_6999198 | 0.04 |
ENST00000382689.8
|
DEFA1B
|
defensin alpha 1B |
| chr2_-_105396943 | 0.04 |
ENST00000409807.5
|
FHL2
|
four and a half LIM domains 2 |
| chr6_+_29461440 | 0.04 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr20_-_31406207 | 0.04 |
ENST00000376314.3
|
DEFB121
|
defensin beta 121 |
| chr10_-_112446932 | 0.04 |
ENST00000682195.1
ENST00000683505.1 ENST00000683895.1 ENST00000682839.1 ENST00000682055.1 |
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr6_+_132552693 | 0.03 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chr10_+_7703300 | 0.03 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr15_-_34336749 | 0.03 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr7_+_142749465 | 0.03 |
ENST00000486171.5
ENST00000619214.4 ENST00000311737.12 |
PRSS1
|
serine protease 1 |
| chr19_-_36489435 | 0.03 |
ENST00000434377.6
ENST00000424129.7 ENST00000452939.6 ENST00000427002.2 |
ZNF566
|
zinc finger protein 566 |
| chr10_-_112446891 | 0.03 |
ENST00000369404.3
ENST00000369405.7 ENST00000626395.2 |
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr19_-_36489854 | 0.03 |
ENST00000493391.6
|
ZNF566
|
zinc finger protein 566 |
| chr19_-_36489553 | 0.03 |
ENST00000392170.7
|
ZNF566
|
zinc finger protein 566 |
| chr15_+_76336755 | 0.03 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr6_-_24935942 | 0.03 |
ENST00000645100.1
ENST00000643898.2 ENST00000613507.4 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr12_-_102061946 | 0.03 |
ENST00000240079.11
|
WASHC3
|
WASH complex subunit 3 |
| chr10_+_7703340 | 0.03 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_202462730 | 0.03 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr10_-_112446734 | 0.03 |
ENST00000684507.1
|
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr7_-_87059639 | 0.03 |
ENST00000450689.7
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr8_-_7018295 | 0.03 |
ENST00000327857.7
|
DEFA3
|
defensin alpha 3 |
| chr8_+_28338640 | 0.03 |
ENST00000522209.1
|
PNOC
|
prepronociceptin |
| chr13_-_48413105 | 0.03 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr11_-_33869816 | 0.03 |
ENST00000395833.7
|
LMO2
|
LIM domain only 2 |
| chr17_-_41047267 | 0.03 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chrX_-_77895546 | 0.03 |
ENST00000358075.11
|
MAGT1
|
magnesium transporter 1 |
| chr11_-_63144221 | 0.03 |
ENST00000417740.5
ENST00000612278.4 ENST00000326192.5 |
SLC22A24
|
solute carrier family 22 member 24 |
| chr4_+_141636923 | 0.03 |
ENST00000529613.5
|
IL15
|
interleukin 15 |
| chrX_+_1615049 | 0.03 |
ENST00000381241.9
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr7_+_142791635 | 0.03 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr4_-_82844418 | 0.03 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chrX_-_77895414 | 0.03 |
ENST00000618282.5
ENST00000373336.3 |
MAGT1
|
magnesium transporter 1 |
| chr5_-_159099745 | 0.03 |
ENST00000517373.1
|
EBF1
|
EBF transcription factor 1 |
| chr12_+_71686473 | 0.03 |
ENST00000549735.5
|
TMEM19
|
transmembrane protein 19 |
| chr12_-_30735014 | 0.03 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr5_-_159099684 | 0.03 |
ENST00000380654.8
|
EBF1
|
EBF transcription factor 1 |
| chr4_-_69653223 | 0.03 |
ENST00000286604.8
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus |
| chr3_+_119186716 | 0.02 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr12_-_70637405 | 0.02 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr14_-_35121950 | 0.02 |
ENST00000554361.5
ENST00000261475.10 |
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr12_-_102062079 | 0.02 |
ENST00000545679.5
|
WASHC3
|
WASH complex subunit 3 |
| chr9_-_35650902 | 0.02 |
ENST00000259608.8
ENST00000618781.1 |
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr1_-_110519175 | 0.02 |
ENST00000369771.4
|
KCNA10
|
potassium voltage-gated channel subfamily A member 10 |
| chr19_+_926001 | 0.02 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr2_+_102311502 | 0.02 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr7_+_77840122 | 0.02 |
ENST00000450574.5
ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_-_110445540 | 0.02 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr6_-_169250825 | 0.02 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr21_+_34364003 | 0.02 |
ENST00000290310.4
|
KCNE2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr8_+_12108172 | 0.02 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D |
| chr17_-_59707404 | 0.02 |
ENST00000393038.3
|
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr17_-_43125450 | 0.02 |
ENST00000494123.5
ENST00000468300.5 ENST00000471181.7 ENST00000652672.1 |
BRCA1
|
BRCA1 DNA repair associated |
| chr3_+_108822759 | 0.02 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_+_61905646 | 0.02 |
ENST00000311832.5
|
COMMD1
|
copper metabolism domain containing 1 |
| chr3_+_108822778 | 0.02 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_+_55267584 | 0.02 |
ENST00000575345.5
|
HLF
|
HLF transcription factor, PAR bZIP family member |
| chrM_+_10759 | 0.02 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4 |
| chr5_-_159099909 | 0.02 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1 |
| chr2_+_68974573 | 0.02 |
ENST00000673932.3
ENST00000377938.4 |
GKN1
|
gastrokine 1 |
| chr1_-_44141631 | 0.02 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr9_+_104094557 | 0.02 |
ENST00000374787.7
|
SMC2
|
structural maintenance of chromosomes 2 |
| chr2_+_201132872 | 0.02 |
ENST00000470178.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chrM_+_10055 | 0.02 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chr2_+_201132769 | 0.02 |
ENST00000494258.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chrX_+_1615158 | 0.02 |
ENST00000381229.9
ENST00000381233.8 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr5_+_157743703 | 0.02 |
ENST00000286307.6
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr9_+_89318492 | 0.02 |
ENST00000375807.8
ENST00000339901.8 |
SECISBP2
|
SECIS binding protein 2 |
| chr4_-_46389351 | 0.02 |
ENST00000503806.5
ENST00000356504.5 ENST00000514090.5 ENST00000506961.5 |
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr2_+_165294031 | 0.02 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr11_-_26722051 | 0.02 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chrX_-_140784366 | 0.02 |
ENST00000674533.1
|
CDR1
|
cerebellar degeneration related protein 1 |
| chr8_+_24440930 | 0.02 |
ENST00000441335.6
ENST00000175238.10 ENST00000380789.5 |
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr6_+_118548289 | 0.02 |
ENST00000357525.6
|
PLN
|
phospholamban |
| chr16_+_56451513 | 0.02 |
ENST00000562150.5
ENST00000561646.5 ENST00000566157.6 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron dependent oxygenase domain containing 1 |
| chr5_-_148654522 | 0.02 |
ENST00000377888.8
|
HTR4
|
5-hydroxytryptamine receptor 4 |
| chr3_-_108058361 | 0.02 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr11_+_56176618 | 0.02 |
ENST00000312298.1
|
OR5J2
|
olfactory receptor family 5 subfamily J member 2 |
| chr2_-_208190001 | 0.02 |
ENST00000451346.5
ENST00000341287.9 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr14_+_61697622 | 0.02 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr10_+_35195124 | 0.02 |
ENST00000487763.5
ENST00000473940.5 ENST00000488328.5 ENST00000356917.9 |
CREM
|
cAMP responsive element modulator |
| chr5_-_154938181 | 0.02 |
ENST00000285873.8
|
GEMIN5
|
gem nuclear organelle associated protein 5 |
| chr7_+_136868622 | 0.02 |
ENST00000680005.1
ENST00000445907.6 |
CHRM2
|
cholinergic receptor muscarinic 2 |
| chr14_-_106538331 | 0.02 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr3_+_172039556 | 0.01 |
ENST00000415807.7
ENST00000421757.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr6_-_89819699 | 0.01 |
ENST00000439638.1
ENST00000629399.2 ENST00000369393.8 |
MDN1
|
midasin AAA ATPase 1 |
| chr11_+_101914997 | 0.01 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chr2_-_135837170 | 0.01 |
ENST00000264162.7
|
LCT
|
lactase |
| chr20_-_50958520 | 0.01 |
ENST00000371588.10
ENST00000371584.9 ENST00000413082.1 ENST00000371582.8 |
DPM1
|
dolichyl-phosphate mannosyltransferase subunit 1, catalytic |
| chr6_-_159044980 | 0.01 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr3_+_98147479 | 0.01 |
ENST00000641380.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr18_-_72865680 | 0.01 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr11_+_122862303 | 0.01 |
ENST00000533709.1
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr4_-_163332589 | 0.01 |
ENST00000296533.3
ENST00000509586.5 ENST00000504391.5 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr18_+_13612614 | 0.01 |
ENST00000586765.1
ENST00000677910.1 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr18_+_13611764 | 0.01 |
ENST00000585931.5
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr10_+_88664439 | 0.01 |
ENST00000394375.7
ENST00000608620.5 ENST00000238983.9 ENST00000355843.2 |
LIPF
|
lipase F, gastric type |
| chrX_+_78945332 | 0.01 |
ENST00000544091.1
ENST00000171757.3 |
P2RY10
|
P2Y receptor family member 10 |
| chr20_-_45579277 | 0.01 |
ENST00000289953.3
|
WFDC8
|
WAP four-disulfide core domain 8 |
| chr2_+_169584385 | 0.01 |
ENST00000462903.6
ENST00000676756.1 |
PPIG
|
peptidylprolyl isomerase G |
| chr18_+_13611910 | 0.01 |
ENST00000590308.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr6_-_134950081 | 0.01 |
ENST00000367847.2
ENST00000265605.7 ENST00000367845.6 |
ALDH8A1
|
aldehyde dehydrogenase 8 family member A1 |
| chr20_+_45629717 | 0.01 |
ENST00000372643.4
|
WFDC10A
|
WAP four-disulfide core domain 10A |
| chr8_-_78805306 | 0.01 |
ENST00000639719.1
ENST00000263851.9 |
IL7
|
interleukin 7 |
| chrX_+_146809768 | 0.01 |
ENST00000438525.3
|
CXorf51B
|
chromosome X open reading frame 51B |
| chr15_+_58138368 | 0.01 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr16_+_25216943 | 0.01 |
ENST00000219660.6
|
AQP8
|
aquaporin 8 |
| chr19_-_51368979 | 0.01 |
ENST00000601435.1
ENST00000291715.5 |
CLDND2
|
claudin domain containing 2 |
| chr9_-_41189310 | 0.01 |
ENST00000456520.5
ENST00000377391.8 ENST00000613716.4 ENST00000617933.1 |
CBWD6
|
COBW domain containing 6 |
| chrX_-_146814726 | 0.01 |
ENST00000458472.1
|
CXorf51A
|
chromosome X open reading frame 51A |
| chr11_+_30231000 | 0.01 |
ENST00000254122.8
ENST00000533718.2 ENST00000417547.1 |
FSHB
|
follicle stimulating hormone subunit beta |
| chr8_-_78805515 | 0.01 |
ENST00000379113.6
ENST00000541183.2 |
IL7
|
interleukin 7 |
| chr6_-_130956371 | 0.01 |
ENST00000639623.1
ENST00000525193.5 ENST00000527659.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr9_+_68241854 | 0.01 |
ENST00000616550.4
ENST00000618217.4 ENST00000377342.9 ENST00000478048.5 ENST00000360171.11 |
CBWD3
|
COBW domain containing 3 |
| chr12_+_62466791 | 0.01 |
ENST00000641654.1
ENST00000546600.5 ENST00000393630.8 ENST00000552738.5 ENST00000393629.6 ENST00000552115.5 |
MON2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr13_+_48232609 | 0.01 |
ENST00000649266.1
|
ITM2B
|
integral membrane protein 2B |
| chr1_-_27155118 | 0.01 |
ENST00000263980.8
|
SLC9A1
|
solute carrier family 9 member A1 |
| chr13_-_108218293 | 0.01 |
ENST00000405925.2
|
LIG4
|
DNA ligase 4 |
| chr20_-_45579326 | 0.01 |
ENST00000357199.8
|
WFDC8
|
WAP four-disulfide core domain 8 |
| chr12_+_57488059 | 0.01 |
ENST00000628866.2
ENST00000262027.10 |
MARS1
|
methionyl-tRNA synthetase 1 |
| chr14_+_45135917 | 0.01 |
ENST00000267430.10
ENST00000556036.5 ENST00000542564.6 |
FANCM
|
FA complementation group M |
| chr6_+_152697934 | 0.01 |
ENST00000532295.1
|
MYCT1
|
MYC target 1 |
| chr19_-_41844629 | 0.01 |
ENST00000609812.6
|
LYPD4
|
LY6/PLAUR domain containing 4 |
| chr2_+_102311546 | 0.01 |
ENST00000233954.6
ENST00000447231.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr13_-_46182136 | 0.01 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr1_+_248231417 | 0.01 |
ENST00000641868.1
|
OR2M4
|
olfactory receptor family 2 subfamily M member 4 |
| chr7_-_30550761 | 0.01 |
ENST00000598361.4
ENST00000440438.6 |
ENSG00000281039.1
GARS1-DT
|
novel protein GARS1 divergent transcript |
| chr9_-_90642791 | 0.01 |
ENST00000375765.5
ENST00000636786.1 |
DIRAS2
|
DIRAS family GTPase 2 |
| chr17_-_42018488 | 0.01 |
ENST00000589773.5
ENST00000674214.1 |
DNAJC7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr2_-_108989206 | 0.01 |
ENST00000258443.7
ENST00000409271.5 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr19_+_35748549 | 0.01 |
ENST00000301159.14
|
LIN37
|
lin-37 DREAM MuvB core complex component |
| chr3_+_138621225 | 0.01 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr17_+_47831608 | 0.01 |
ENST00000269025.9
|
LRRC46
|
leucine rich repeat containing 46 |
| chr1_+_196774813 | 0.01 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr2_-_127884860 | 0.00 |
ENST00000393001.1
|
AMMECR1L
|
AMMECR1 like |
| chr11_-_4697831 | 0.00 |
ENST00000641159.1
ENST00000396950.4 ENST00000532598.1 |
OR51C1P
OR51E2
|
olfactory receptor family 51 subfamily C member 1 pseudogene olfactory receptor family 51 subfamily E member 2 |
| chr6_-_15548360 | 0.00 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr4_-_64409444 | 0.00 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr3_+_138621207 | 0.00 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_-_10292901 | 0.00 |
ENST00000430179.5
ENST00000287656.11 ENST00000437422.6 ENST00000439975.6 ENST00000446937.2 ENST00000449238.6 ENST00000457360.5 |
GHRL
|
ghrelin and obestatin prepropeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.0 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.0 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |