Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
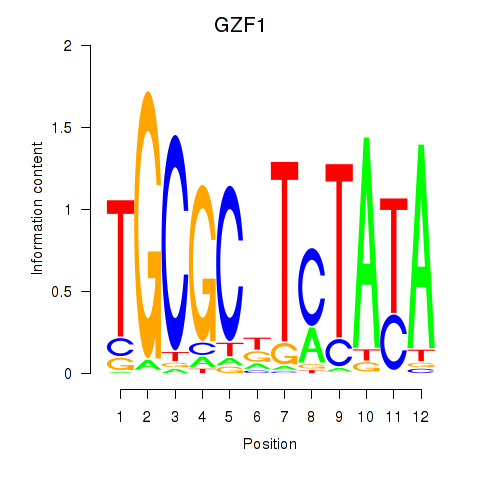
Results for GZF1
Z-value: 0.17
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.16 | GDNF inducible zinc finger protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GZF1 | hg38_v1_chr20_+_23362144_23362320 | 0.14 | 7.5e-01 | Click! |
Activity profile of GZF1 motif
Sorted Z-values of GZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_33080445 | 0.23 |
ENST00000428835.5
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr3_-_86991135 | 0.16 |
ENST00000398399.7
|
VGLL3
|
vestigial like family member 3 |
| chr1_+_78649818 | 0.15 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr1_+_148889403 | 0.14 |
ENST00000464103.5
ENST00000534536.5 ENST00000369356.8 ENST00000369354.7 ENST00000369347.8 ENST00000369349.7 ENST00000369351.7 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr7_-_27143672 | 0.14 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr14_-_52069039 | 0.14 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
| chr14_-_52069228 | 0.13 |
ENST00000617139.4
|
NID2
|
nidogen 2 |
| chr6_-_33080710 | 0.12 |
ENST00000419277.5
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chrX_+_54809060 | 0.11 |
ENST00000396224.1
|
MAGED2
|
MAGE family member D2 |
| chr1_+_26280117 | 0.10 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamate rich protein like 3 |
| chr19_-_43880142 | 0.10 |
ENST00000324394.7
|
ZNF404
|
zinc finger protein 404 |
| chr1_+_26280059 | 0.09 |
ENST00000270792.10
|
SH3BGRL3
|
SH3 domain binding glutamate rich protein like 3 |
| chrX_+_81202066 | 0.09 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chr19_-_36054224 | 0.08 |
ENST00000292894.2
|
THAP8
|
THAP domain containing 8 |
| chrX_+_103628692 | 0.08 |
ENST00000372626.7
|
TCEAL1
|
transcription elongation factor A like 1 |
| chr11_-_17476801 | 0.06 |
ENST00000644772.1
ENST00000642271.1 ENST00000684571.1 ENST00000646902.1 ENST00000647015.1 ENST00000302539.9 ENST00000389817.8 ENST00000643260.1 ENST00000683136.1 |
ABCC8
|
ATP binding cassette subfamily C member 8 |
| chr19_+_12995467 | 0.06 |
ENST00000592199.6
|
NFIX
|
nuclear factor I X |
| chr6_+_30557274 | 0.06 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr6_+_30557287 | 0.05 |
ENST00000376560.8
|
PRR3
|
proline rich 3 |
| chr4_+_26343156 | 0.05 |
ENST00000680928.1
ENST00000681260.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_+_86578543 | 0.05 |
ENST00000320241.5
|
FOXL1
|
forkhead box L1 |
| chr16_-_1611985 | 0.05 |
ENST00000426508.7
|
IFT140
|
intraflagellar transport 140 |
| chr7_-_26864573 | 0.04 |
ENST00000345317.7
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr19_+_926001 | 0.04 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr12_+_106774630 | 0.04 |
ENST00000392839.6
ENST00000548914.5 ENST00000355478.6 ENST00000552619.1 ENST00000549643.5 ENST00000392837.9 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr11_+_65572349 | 0.04 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.2 |
FAM89B
|
family with sequence similarity 89 member B |
| chr12_+_104064520 | 0.04 |
ENST00000229330.9
|
HCFC2
|
host cell factor C2 |
| chrX_+_153972729 | 0.03 |
ENST00000369982.5
|
TMEM187
|
transmembrane protein 187 |
| chr19_-_51630401 | 0.03 |
ENST00000683636.1
|
SIGLEC5
|
sialic acid binding Ig like lectin 5 |
| chr6_-_36547400 | 0.03 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38 |
| chr17_-_38748184 | 0.03 |
ENST00000618941.4
ENST00000620225.5 ENST00000618506.1 ENST00000616129.4 |
PCGF2
|
polycomb group ring finger 2 |
| chr1_-_41918666 | 0.02 |
ENST00000372584.5
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr14_-_59484006 | 0.02 |
ENST00000481608.1
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr14_-_59484317 | 0.02 |
ENST00000247194.9
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr1_-_41918858 | 0.02 |
ENST00000372583.6
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr3_+_10992175 | 0.02 |
ENST00000646702.1
|
SLC6A1
|
solute carrier family 6 member 1 |
| chr19_+_38390055 | 0.02 |
ENST00000587947.5
ENST00000338502.8 |
SPRED3
|
sprouty related EVH1 domain containing 3 |
| chr1_-_32336224 | 0.02 |
ENST00000329421.8
|
MARCKSL1
|
MARCKS like 1 |
| chr19_-_51646800 | 0.02 |
ENST00000599649.5
ENST00000429354.3 ENST00000360844.6 |
SIGLEC5
SIGLEC14
|
sialic acid binding Ig like lectin 5 sialic acid binding Ig like lectin 14 |
| chr19_-_58573555 | 0.02 |
ENST00000599369.5
|
MZF1
|
myeloid zinc finger 1 |
| chr11_+_102047422 | 0.01 |
ENST00000434758.7
ENST00000526781.5 ENST00000534360.1 |
CFAP300
|
cilia and flagella associated protein 300 |
| chr19_-_58573280 | 0.01 |
ENST00000594234.5
ENST00000596039.1 ENST00000215057.7 |
MZF1
|
myeloid zinc finger 1 |
| chr20_-_32743406 | 0.01 |
ENST00000474815.2
ENST00000446419.6 ENST00000278980.11 ENST00000642484.1 ENST00000646357.1 |
COMMD7
ENSG00000285382.1
|
COMM domain containing 7 novel protein |
| chr9_+_134326435 | 0.01 |
ENST00000481739.2
|
RXRA
|
retinoid X receptor alpha |
| chr1_-_226186673 | 0.01 |
ENST00000366812.6
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr9_-_14307928 | 0.01 |
ENST00000637640.1
ENST00000493697.1 ENST00000636057.1 |
NFIB
|
nuclear factor I B |
| chr7_+_5045821 | 0.01 |
ENST00000353796.7
ENST00000396912.2 ENST00000396904.2 |
RBAK
RBAK-RBAKDN
|
RB associated KRAB zinc finger RBAK-RBAKDN readthrough |
| chr22_-_42959852 | 0.01 |
ENST00000402229.5
ENST00000407585.5 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr16_+_67842277 | 0.01 |
ENST00000303596.3
|
THAP11
|
THAP domain containing 11 |
| chr1_-_2530256 | 0.01 |
ENST00000378453.4
|
HES5
|
hes family bHLH transcription factor 5 |
| chr9_+_128371343 | 0.01 |
ENST00000372850.5
ENST00000372847.1 ENST00000372853.9 ENST00000483206.2 |
URM1
|
ubiquitin related modifier 1 |
| chr2_-_149587602 | 0.01 |
ENST00000428879.5
ENST00000303319.10 ENST00000422782.2 |
MMADHC
|
metabolism of cobalamin associated D |
| chr1_-_120051714 | 0.00 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr7_-_6706843 | 0.00 |
ENST00000394917.3
ENST00000342651.9 ENST00000405858.6 |
ZNF12
|
zinc finger protein 12 |
| chr19_-_10315987 | 0.00 |
ENST00000393708.3
ENST00000494368.5 |
FDX2
|
ferredoxin 2 |
| chr1_+_40709475 | 0.00 |
ENST00000372651.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of GZF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:1905073 | positive regulation of relaxation of muscle(GO:1901079) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.0 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |